Figure 1.
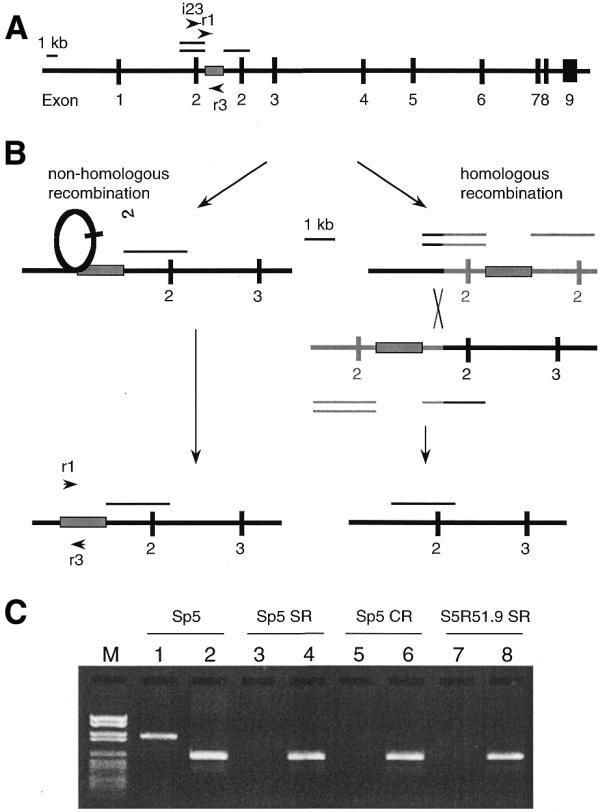
Molecular mechanisms for reversion of the hprt gene in Sp5 cells to wild-type. (A) Duplication of exon 2 in the hprt gene of the Sp5 cell line results in a non-functional gene, which can be selected for on the basis of its [HPRT]– phenotype. The single line designates the parental region, while the double lines indicate the duplicated copy of exon 2 together with flanking intron regions (24,25). The box represents the sequence between the duplicated regions. Arrowheads indicate the primers employed. (B) Recombination pathways for reversion to a [HPRT]+ phenotype: if the reversion from a [HPRT]– to a [HPRT]+ phenotype involves non-homologous recombination, the region represented by the box will be retained in the revertants. If homologous recombination is involved in this reversion event, this region will invariably be lost. (C) Lanes 1, 3, 5 and 7 contain the PCR products obtained employing the primers i23 and r3 together with DNA from Sp5 cells or from spontaneous (Sp5 SR), CPT-induced (Sp5 CR) Sp5 revertants or from spontaneous S5R51.9 revertants (S5R51.9 SR), respectively. This gel demonstrates that the duplicated exon 2 has been lost in all of these revertants. Lanes 2, 4, 6 and 8 contain the PCR products obtained employing the primers r1 and r3 together with DNA from Sp5 cells or from Sp5 SR, Sp5 CR or S5R51.9 SR, respectively, and demonstrate that the DNA region (boxed) is present in all of these revertants, i.e., that non-homologous recombination was involved in restoring the functional hprt gene. Identical results were obtained for all of the 32 revertants analysed. Marker VI (Boehringer Mannheim) was used as a size indicator. Thus, these data demonstrate that, in all revertants, the original duplication of exon 2 is lost, whereas the DNA region originally located between the duplicated regions (indicated by the box) is retained.
