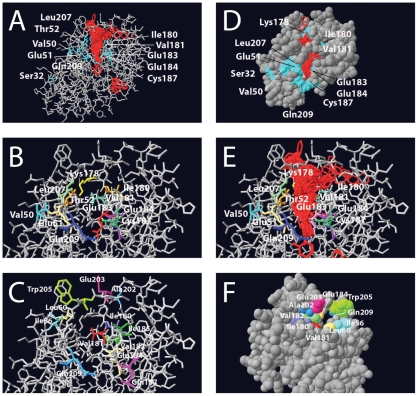
Figure 2. Details of the modeled tertiary structure of the HbpR A-domain, showing amino acid residues that were mutated in this study and the region onto which 2-HBP is predicted to be bound.
(A) Results of 1000 iterations of 2-HBP (in red) docking calculations using gramm onto the predicted HbpR A-domain protein surface, whilst indicating the position of residues altered to Phe. (B) Close-up of the same, but without the docked 2-HBP positions. (C) as for B, now highlighting the other changed residues. (D) Van der Waals-filled model slightly turned compared to A, in order to indicate the region of 2-HBP docked molecules. (E), as B, but with 2-HBP docked positions. (F) Turned van der Waals-filled model showing the tunnel from the other side of the entry.