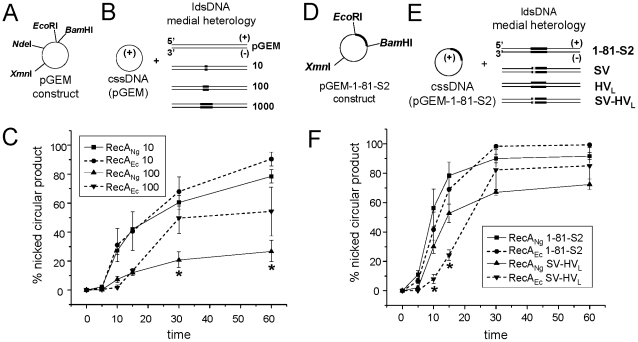
Figure 6. Strand exchange with DNA substrates to mimic DNA transformation and antigenic variation in vitro.
A. Schematic of pGEM vector with relevant restriction sites used to clone heterologous inserts (see Materials and Methods). B. Linear dsDNA heterologous inserts digested with NdeI to give medial heterology and reacted with pGEM cssDNA. C. % nicked circular product observed in strand exchange reactions promoted by RecANg and RecAEc using pGEM circular ssDNA reacted with pGEM-10 and pGEM-100 linear dsDNA (designated “10” and “100” in Figure). Error bars represent the standard error of the mean of 3 independent experiments. *P<0.05 by Students two-tailed t-test (RecANg 100 compared to RecAEc 100). D. Schematic of construct pGEM 1-81-S2 containing the 1-81-S2 pilE DNA sequence and relevant restriction sites. E. Linear dsDNA of antigenic variants SV, HVL, and SV-HVL heterologies (designated with shading and cross-hatches, see Figure S3 and Materials and Methods) digested with XmnI to give medial heterology and reacted with pGEM 1-81-S2 circular ssDNA. F. % nicked circular product observed in strand exchange reactions promoted by RecAEc and RecANg using pGEM-1-81-S2 cssDNA reacted with pGEM-1-81-S2 or pGEM-SV-HVL linear dsDNA. Error bars represent the standard error of the mean of at least 3 independent experiments. *P<0.05 by Students two-tailed t-test (RecANg SV-HVL relative to RecAEc SV-HVL).