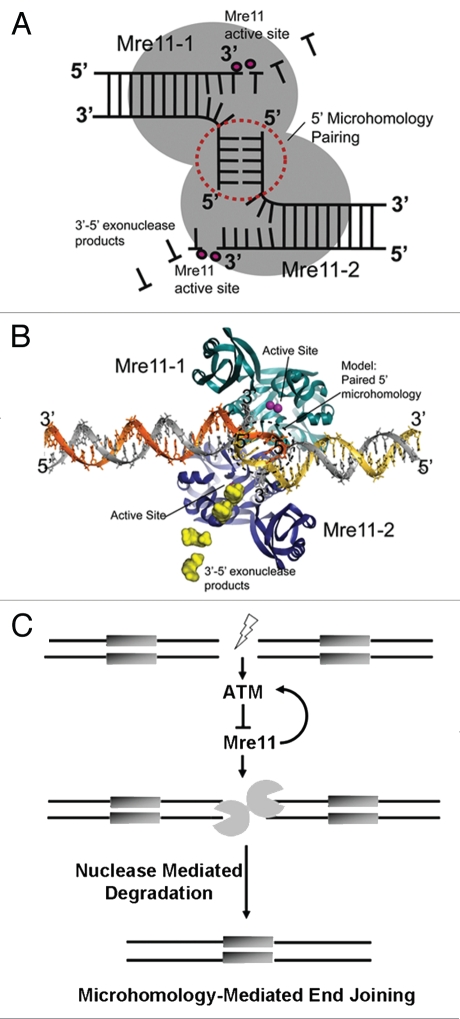
Figure 8.
Mre11-mediated microhomology-mediated end-joining and its regulation by ATM. (A) Schematic for Mre11 dimer-mediated MMEJ. The Mre11 homodimer is a symmetric structure that binds two DNA ends. 5′ termini are paired at the interface of the dimer, whereas 3′ ends are degraded by Mre11 3′–5′ exonuclease activity in Mre11 active site (B) Structure-based model for an Mre11 5′-microhomology end joining complex based on the Mre11-DNA synaptic complex (PDB ID 3DSC). The 5 bp 5′ microhomology (circled dotted line) was modeled by extending the 5′ termini of DNAs observed in the X-ray structure. 5′ MMEJ pairing could occur in the space between crystallographically defined DNA interaction sites. (C) Model delineating the role of ATM in suppression of error-prone DSB repair via microhomology-mediated end joining. ATM is activated upon the formation of a DSB. Then, its kinase activity maintains end-stability at a break through suppression of Mre11-dependent microhomology-directed DNA degradation and subsequent end joining.