Abstract
We utilized gene expression profiling of a comprehensive panel of purified developmentally defined normal murine B cells to identify unique transcriptional signatures for each subset. To elucidate transcription factor activities that function in a stage-specific fashion, we used gene sets that share transcription factor targets and found that germinal center B cells had a robust enrichment of up-regulated and down-regulated signatures compared with the other B-cell subsets. Notably, we found Yy1 and its targets to be central regulators of the germinal center B (GCB)-specific transcriptional program with binding of Yy1 to select signature genes in GCB cells, and translation of the Yy1 signatures to human GCB cells. We then tested whether our newly generated, stage-specific transcriptional signatures could be used to link murine lymphoma models to stages of normal B-cell development. Although each of the molecularly defined murine lymphoma models conserved certain stage-specific features of normal B-cell development, there was a significant alteration of the normal differentiation signature following malignant transformation. These findings offer important tools and insights for elucidating differences between normal and malignant B cells.
B-cell maturation proceeds through distinct developmental stages that are largely characterized by the generation and modification of antibodies that function during adaptive immune responses (1–3). This hierarchical development is controlled in part by the activation/inactivation of transcription factors (TF), resulting in the induction or suppression of large sets of genes that are necessary for the cellular processes specific to discrete states or for further differentiation (4). Therefore, by understanding the genes that are expressed with specificity for given differentiation states, we can gain insight into the TFs that are central to discrete B-cell programs. Germinal center B (GCB) cells are of particular interest because of their importance in increasing the efficacy and diversity of adaptive immune responses via directed DNA modifications [somatic hypermutation (SHM)] and double-stranded breaks [class-switch recombination (CSR)]. Errors in the programmed DNA alterations inherent to GCB cells can lead to translocations and aberrant SHM that are associated with B-cell lymphoma. Although certain GCB-specific transcriptional regulators have been defined (5), much remains to be characterized in this highly dynamic B-cell subset.
One approach for gaining insight into mechanisms of B-cell development and survival has been to generate specific genetic mutations in murine genes modeling those found in human B-cell lymphoma. For genes with functions in controlling B-cell development or survival, this can result in alterations of B-cell homeostasis, and can occasionally promote lymphomagenesis. Murine lymphoma models can be putatively aligned with normal stages of B-cell differentiation by using the expression of surface markers and the status of Immunoglobulin (Ig) gene segment rearrangements, notably V(D)J or CSR Ig gene rearrangement status and SHM. These comparisons provide important insights with regard to the gene function; but the degree to which these mutant models recapitulate normal B-cell biology, and whether the similarities or differences are the more important features, remains unknown.
Each discrete B-cell differentiation state can be defined by cell surface antigen profiles that facilitate classification. However, few studies have applied modern molecular profiling tools to interrogate panels of B cells from all differentiation states (6). Furthermore, many of these studies have been focused on early differentiation states associated with V(D)J recombination (7, 8). Here, we used transcriptional profiling of a comprehensive panel of normal B cells at each of the major developmental stages to gain insight into the relative differences in their signatures and to infer relative TF activities.
Because of the multiple mechanisms of posttranscriptional regulation of TF activity, and the inability of TF transcript levels to reflect these modifications, we used bioinformatic analysis of TF target gene sets and conserved DNA sequence motifs to interrogate TF activity. With this approach, we identified Yy1 as a likely central regulator of the GCB-specific transcriptional program. Binding of Yy1 to GCB cell signature genes was confirmed by ChIP, and Yy1 target genes were coordinately expressed in human GCB cells.
In addition, we used our comprehensive transcriptional signatures to interrogate selected murine lymphoma models that putatively represent selected normal B-cell differentiation states, and found that a small, but significant portion of normal B-cell signatures are reconstituted in malignant murine B cells. Together, these findings not only provide a resource of transcriptional signatures of murine B-cell development, but highlight the utility of such signatures in identifying central regulators of B-cell development and in gaining insight into the relationship between normal and malignant B cells.
Results and Discussion
Purification and Transcriptional Profiling of Murine B-Cell Subsets.
Most classification schemes for B-cell development have chiefly relied on the DNA rearrangement status of the Ig receptor gene segments, together with the expression pattern of the BCR and/or specific surface markers to define discrete stages. Although this is an extremely practical classification system, dynamic changes that occur at specific developmental stages are not readily illustrated. Global gene expression profiling has provided many insights into immunological processes (9), and recent developments in bioinformatic approaches have allowed gene expression signatures to be used to infer biological processes such as transcription factor activity (10).
To profile discrete B-cell differentiation states, we used previously defined immunophenotypic characteristics to define and purify each subtype from the bone marrow and spleen of mice by FACS (Table 1). B-cell subtypes included pro-, pre-, transitional (Trans.), follicular (Foll.), marginal zone (MZ), and germinal center B cells (GCB), as well as plasmablasts (P.blasts) and plasma cells (P. Cells) that were >90% pure. These samples were subsequently used for global transcriptional profiling on Affymetrix Mouse 430 A2 microarrays. Using known B-cell differentiation antigens (11–13), we first confirmed that markers were expressed in the appropriate B-cell subsets, including Dntt, Sox4, and Rag1 in pro- and pre-B cells, Aicda and Fas in GCB, and Xbp1 in P.blasts (Fig. S1).
Table 1.
Origin, immunophenotypic characteristics and immunization status of B-cell differentiation subsets
| Subset | Origin | Surface markers (defined for sort) | Immune status |
| Pro-B | BM | B220+/Cd43high/IgMneg | Naive |
| Pre-B | BM | B220+/Cd43low/IgMneg | Naive |
| Trans. | Spleen | B220+/Cd93+ | Naive |
| Foll. B | Spleen | Cd19+/Cd21intermediate/Cd23high | Naive |
| MZ | Spleen | Cd19+/Cd21high/Cd23low | Naive |
| GCB | Spleen | B220+/PNAhigh | Immunized |
| P. blast | Spleen | B220+/Cd138+/Cd93− | Immunized |
| P. cell | BM | B220low/Cd138+/Cd93− | Immunized |
BM, bone marrow.
Comprehensive Transcriptional Profiling of B-Cell Subsets Highlights Key Transcriptional Regulators.
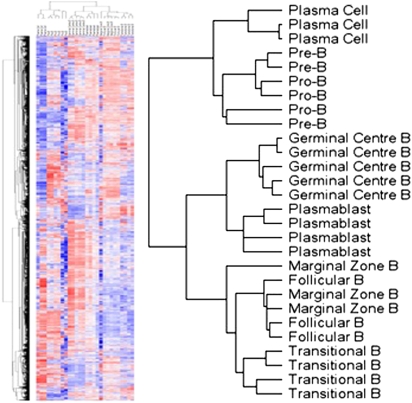
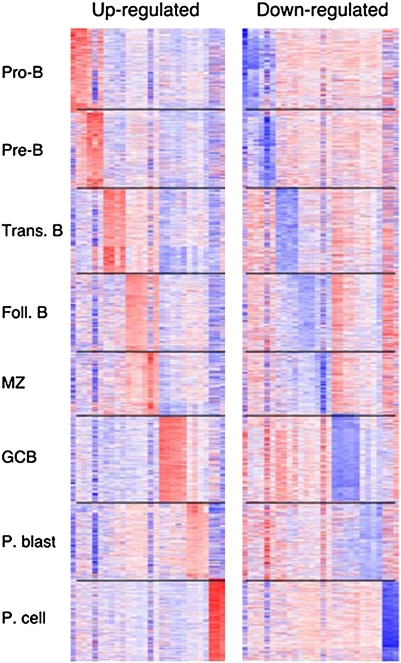
We performed unsupervised hierarchical clustering within the space of the top 3,000 markers as ranked by median absolute deviation and largely confirmed the cosegregation of samples assigned to the same subset, although the Pro-B and Pre-B samples and the Foll. and MZ samples were less divergent (Fig. 1). Given the subtle differences in the immunophenotypes of Pro- and Pre-B cells and of Foll. and MZ B cells (Table 1), the transcriptional similarities were expected. We next used the transcriptional profiles and a supervised one-versus-all analysis to define signatures that were specific to each B-cell differentiation state. These signatures, 350 up-regulated and 350 down-regulated genes that were specifically and significantly [False Discovery Rate Q-value (FDR) < 0.25] altered in transcript abundance within each B-cell subset, are shown and listed in Fig. 2 and Dataset S1, respectively. Despite each gene being significantly over- or underexpressed in individual subsets compared with all remaining subsets, the more subtle distinctions between Pro-B and Pre-B subsets and between Foll. and MZ subsets are apparent (Fig. 2). In contrast, the GCB signatures are highly specific, indicating that the signature genes may be useful for gaining insight into GCB-specific biologic processes.
Fig. 1.
Unsupervised clustering of B-cell subsets. Unsupervised hierarchical clustering of individual samples within the space of the 3,000 most variably expressed genes (Left) resulted in cosegregation of samples with those of the same or similar differentiation state, as shown by the enlargement of the clustering dendogram (Right).
Fig. 2.
Distinct transcriptional signatures of B-cell subsets. Signatures consisting of 350 significantly (FDR < 0.25) up-regulated and 350 significantly down-regulated genes were defined and are shown for each subset.
In addition to including known B-cell subtype markers such as Dntt in Pro-B cells, Sox4 in Pro- and Pre-B cells, Irf4 in Trans. B cells, and Fas and Aicda in GCB cells, these developmental signatures also extend beyond known markers and include other functionally important genes. For example, GCB cells show specific up-regulation of an important Bcl-2 family member (Mcl-1) (14), a regulator of Polycomb Repressor Complex 2 function (Suz12) (15), and a central regulator of autophagy (Becn1) (16). In addition, these comprehensive stage-specific transcriptional signatures provide a framework for defining transcriptional regulators with selective activity in specific B-cell subsets.
Because of the multiple mechanisms by which TF activity can be modulated at a posttranslational level, the transcript abundance of TFs is not always an accurate indication of their activity (17). For these reasons, we used the transcript abundance of sets of TF target genes to infer activity of specific TFs. The compendium of TF target gene sets used in this investigation was assembled by mapping evolutionarily conserved regions within mammalian genomes and identifying nondegenerate TF binding-sequence motifs within them (18). Because of the lack of defined binding sequence matrices for some TFs and the variable rate of binding degeneracy of others, this approach resulted in some TFs not being having representative target gene sets and other TFs having multiple target gene sets. Our current analysis was restricted to transcription factor targets with well-conserved TF binding sites and multiple target gene sets; certain essential GCB TFs (e.g., Bcl-6) were not represented in these target gene sets.
We used hypergeometic gene set enrichment analysis (HG-GSEA) to define the probability of overlaps between predefined gene sets in a fashion that corrects for multiple hypothesis testing, providing FDR values. Using our B-cell subset-specific transcriptional profiles, we identified significant (FDR < 0.25) enrichment of 251 TF target gene sets within up-regulated signatures and 155 TF target gene sets within down-regulated signatures across all B-cell subsets (Dataset S2). To identify TF target gene sets with the most robust enrichment, we required >50% of target gene sets for each transcription factor to have statistical significance (FDR < 0.25), and ≥1 to be highly significant (FDR < 0.01). Using these stringent criteria, NFκB target genes were significantly up-regulated in Transitional B cells and down-regulated in Pro-B and Pre-B cells (Fig. S2). The identification of NFκB target gene expression in Trans. B cells is in line with previous studies (19, 20) and suggests a stage-specific role for NFκB in early B-cell development.
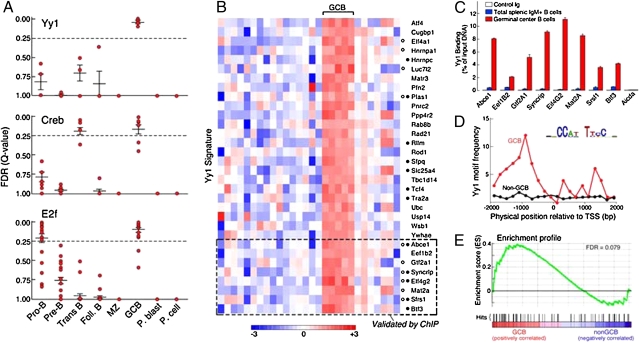
We next used the same rigorous criteria to highlight the most robustly and consistently implicated transcriptional regulators of the GCB signature, and identified three factors as important for the GCB-specific transcriptional program: Yy1, Creb, and E2f (Fig. 3A). Creb gene sets also showed enrichment within the Trans. B-cell subset (Fig. 3A and Fig. S3 A–C), and E2f gene sets also showed significant enrichment in the Pro-B subsets (Fig. 3A and Fig. S3 D–F), suggesting dynamic Creb and E2f activity at specific stages of B-cell development. Creb is of interest in GCB cells because of its previously defined role in promoting B-cell survival in response to activation by BCR and CD40 ligation (21, 22). In addition, E2f promotes proliferation downstream from PI3K/Akt signaling, a central survival pathway (23).
Fig. 3.
Signatures implicate Yy1 as a GCB-specific transcriptional regulator. (A) HG-GSEA of the GCB-specific signature and the enrichment of target gene sets for Yy1 (Top), Creb (Middle), and E2f (Bottom). Each point represents the FDR of an independent gene set within a specific B-cell subset signature. (B) GCB-specific genes within Yy1 target gene sets. Creb and E2F target gene sets are indicated by gray and black circles to the left of gene symbols, respectively. The subset of Yy1 target genes validated by ChIP-qPCR are indicated at Bottom. (C) ChIP-qPCR of GCB-specific Yy1 target genes. (D) Distribution frequency of the Yy1 binding-sequence motif (Top Right) enriched in the GCB signature. (E) GSEA of the 125 murine GCB-specific Yy1 target genes (identified by motif analysis) in human GCB vs. non–GCB-cell transcriptional profiles.
Of the gene sets within our database, Yy1 was the only TF that was specifically enriched in the GCB signature (Fig. 3A). In addition to functioning as a transcriptional activator (24), Yy1 participates in Ig recombination by facilitating IgH locus contraction (25); Yy1 also serves in the recruitment of the polycomb repressor 2 complex (26), which is specifically up-regulated during the germinal center reaction (27). Interestingly, there were a significant number of shared Yy1, Creb, and E2f targets identified in the GCB signature, with 12 of the 33 Yy1 target genes also present in Creb target gene sets, 10 in E2f target gene sets, and five in both Creb and E2f target gene sets (Fig. 3B). In line with previous observations (28, 29), these data suggest that Yy1 may function in regulating the GCB transcriptional program via interaction with certain Creb and E2f factors and modulating the specificity of these factors in the germinal center (Fig. S3).
Yy1 as a Regulator of GCB Signature Genes.
The target gene sets for Yy1 used in HG-GSEA analysis were derived by identifying highly conserved Yy1 DNA binding sequence motifs in multiple species, including mice and humans (18). We first validated Yy1 binding to GCB-specific Yy1 target genes using publicly available ChIP-sequencing data (30) from human K562 cells, and found that >90% (30/33) of predicted GCB-specific Yy1 target genes contained Yy1 binding peaks (Fig. S4). Next, we performed ChIP-qPCR in GCBs and total splenic IgM+ B cells and measured Yy1 binding to eight GCB-specific Yy1 target genes and one GCB-specific signature gene that was not predicted to be a Yy1 target, Aicda. This showed highly significant increases in the binding of Yy1 to predicted target genes in GCBs compared with total splenic IgM+ B cells (mostly Foll. and MZ B cells), and no detectable binding of Yy1 to Aicda (Fig. 3C). Together, these results demonstrate that Yy1 binds to predicted target genes that are selectively expressed in GCBs and validates the computational identification of Yy1 as a key regulator of the GCB transcriptional program.
In addition to identifying Yy1 target genes on the basis of their highly conserved Yy1 binding sites, we directly interrogated GCB signature genes for Yy1 binding sites with a second unbiased approach. GCB signature genes were screened for significantly overrepresented DNA sequence motifs (31). This revealed the presence of 754 significantly enriched (FDR < 0.1) motifs, corresponding to 254 highly-correlated motif clusters. Alignment of clusters to known TF matrices revealed matches to 34 known TF matrices including ubiquitous TFs; of note, one motif cluster was tightly correlated with the Yy1 binding matrix. We therefore reassessed the GCB signature genes and genes of non-GCB signatures for the frequency of the identified Yy1 binding sequence motif in regulatory regions, and identified binding sites in 35% (125/350) of GCB signature genes (Dataset S3), with a peak of Yy1 binding site frequency ∼1 kb upstream of the transcription start site (TSS) of GCB signature genes (Fig. 3D). In contrast, no peak was identified among non-GCB signature genes (Fig. 3D). These results reinforce the specificity of Yy1 for GCB signature genes, and indicate that Yy1 may regulate a larger set of GCB-specific genes than that highlighted by the HG-GSEA analysis.
Given the evidence supporting Yy1 as a key factor in GCB biology, we next asked whether the 125 murine GCB-specific genes possessing Yy1 motifs were selectively overrepresented in human GCB cell profiles. GSEA revealed there to be a significant enrichment of the murine GCB-specific Yy1 target genes within human GCB cells (compared with naive and memory B cells; FDR = 0.079) (Fig. 3E). Taken together, these data suggest that Yy1 is also a key transcriptional regulator of human GCBs.
Normal Differentiation State Signatures in Murine B-Cell Tumor Models.
Murine models are commonly used to gain insight into the role of dysregulated genes in aberrant cell survival, perturbed differentiation, and lymphomagenesis. In certain murine lymphoma models, the malignant B cells are associated with specific stages of normal B-cell differentiation based on the status of Ig genes and the expression of cell surface markers. However, the degree to which these lymphoma cells recapitulate or diverge from a normal B-cell program after transformation remains unknown. To address this question, we used our comprehensive transcriptional signatures of normal B-cell differentiation states to interrogate three lymphoma models that are thought to retain features associated with specific normal stages of differentiation; a double-knockout of the Lig4 and p53 genes (Lig4/p53), a Bcl6 transgenic (IμHABCL6) and a double transgenic of Bcl6 and Myc (IμHABcl6//λMyc). Previous analyses of these three murine tumor models correlated Lig4/p53 lymphomas with the Pro-B/Pre-B-cell stage of differentiation by immunophenotype and the presence of IgH rearrangements (32), IμHABcl6 tumors with the GCB stage by immunophenotype and the presence of SHM (33), and IμHABcl6/λMyc lymphomas with the plasmablastic stage by immunophenotype (34). Interrogation of the transcriptional profiles of these models therefore provided an opportunity to assess the extent to which the malignant lymphomas retained normal B-cell subset signatures. We first evaluated these tumors within the space of the a priori set of known B-cell differentiation markers, and found extensive differences in the patterns of these markers between the tumor models and their putative corresponding normal differentiation states (Fig. S1). For example, Lig4/p53 lymphomas retained expression of the Pro/Pre-B markers Cd93 and Rag1, but not Dntt or Kit. IμHABCL6 tumors maintained expression of GCB genes, Fas, and Mcl1, but not Gabpa (34). IμHABCL6/λMyc lymphomas maintained expression of the P.blast marker Prdm1, but showed variable expression of Xbp1.
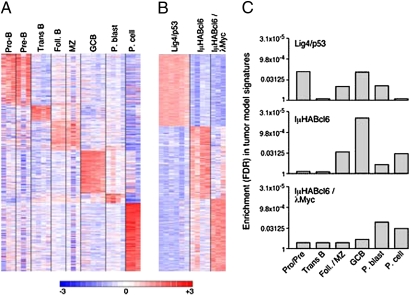
We then asked whether the more comprehensive signatures of normal B-cell subsets could be used to derive a robust classification schema for interrogation of murine lymphoma models (Fig. 4A). For this analysis, we grouped the transcriptionally similar normal B-cell subsets (Pro-B/Pre-B and MZ/Foll.) into single classification categories and assigned an additional filtering criterion to our signature genes (fold change >1.25; Fig. 4A). Tumor model-specific signatures were derived using a one-versus-all analysis, which defined the top 350 genes that were significantly more highly expressed in each model compared with the remaining two models (Fig. 4B). We then evaluated the representation of normal B-cell subset classification signatures within lymphoma model signatures by HG-GSEA. This revealed that the Lig4/p53 knock-out tumors shared components of the normal Pro-B/Pre-B-cell signatures, IμHABcl6 tumors retained features of normal GCB cells, and IμHABcl6/λMyc dual transgenic lymphomas shared more features of normal P.blasts (Fig. 4C). Therefore, there were conserved, stage-specific features in each of these molecularly defined murine lymphoma models. However, the tumors retained only a minor component of the normal B-cell signatures, indicating that the majority of the normal differentiation signature was lost following malignant transformation.
Fig. 4.
Enrichment of normal B-cell signatures in murine lymphoma models. (A) Classification signatures of the normal murine B-cell subsets. (B) Transcriptional signatures of the murine tumor models. (C) HG-GSEA of normal B-cell classification signatures (A) within tumor model signatures (B). The enrichment of the signatures of normal B-cell subsets with which the tumor models are putatively aligned is shown.
Conclusion
Herein, we have undertaken comprehensive transcriptional profiling of a wide developmental range of normal and malignant murine B cells. We have defined robust stage-specific transcriptional signatures of normal murine B-cell differentiation that extend beyond previously identified markers and provide new functional insights. Using these signatures in tandem with HG-GSEA, motif conservation analyses and validation by ChIP, we identified Yy1 as a murine GCB-specific transcriptional regulator. Our analysis used a limited number of transcription factor gene sets based on highly conserved sequences of TF binding sites; additional important factors are not included in the current database. Nevertheless, this approach offers an important tool to interrogate functional transcription factor pathways in B cells, and identifies Yy1 as a member of the set active in GCB cells. Yy1 target genes were also preferentially expressed in human GCB cells. These findings have major implications regarding the potential role of Yy1 in locus contraction during CSR in the GC reaction. We also used the stage-specific normal B-cell signatures to gain insight into the conservation of normal B-cell transcriptional programs in murine lymphoma models. The differentiation signatures could be used to link specific lymphoid tumors with stages of development, although the normal B-cell profiles were largely lost or altered by malignant transformation. This study therefore highlights comprehensive signatures of murine B-cell differentiation states, and provides examples of how these signatures can be applied to gain important functional insights into normal and malignant B cells.
Materials and Methods
B-Cell Isolation and Transcriptional Profiling.
Normal WT mouse B cells were isolated from 8- to 12-wk-old mice. Single-cell suspensions were obtained from each lymphoid organ, stained with antibodies to surface markers (Table 1), and purified by FACS (Fig. S5). Cell purity was >90% of collected events in all cases, as defined by the sorting criteria of each population. Detailed methods regarding B-cell isolation, sorting, and sample preparation are described in SI Materials and Methods.
Gene Expression Data Normalization and Validation.
Raw probe signal intensities for each sample were scaled by robust multiarray normalization and log2 transformed before analysis (35). Because of the similarities in scatter profiles of B and T lymphocytes, we eliminated the possibility of a confounding effect of T cell contamination by the removal of probes corresponding to a T cell signature before analysis (SI Materials and Methods). The purity and origin of each sample included in this study was assessed through the use of an a priori set of known B-cell differentiation markers obtained from a review of the literature (Dataset S4) (11–13). Unsupervised hierarchical clustering was performed within the space of the top 3,000 probes, as ranked by median absolute deviation, using a correlation distance metric and centroid linking method.
Differential Gene Expression Analysis.
Differential gene expression analyses were performed using the top 10,000 median absolute deviation-ranked probes across the data set. Probes correlating with the subset of interest were ranked according to their signal-to-noise ratio (SNR) as previously described (36). Each analysis was corrected for multiple hypothesis testing, to yield FDR Q values, by completing 10,000 permutations (37). Gene signatures for each subset were restricted to 350 up-regulated and 350 down-regulated transcripts, the maximum number that maintained statistical significance (FDR < 0.25) across all subsets.
Hypergeometric Enrichment Analysis of Transcription Factor Target Gene Sets.
The overrepresentation of TF target gene sets within transcriptional signatures was assessed by a one-tailed Fisher exact test using the Gaussian hypergeometric probability distribution (38), henceforth referred to as hypergeometric gene set enrichment analysis (HG-GSEA). We used HG-GSEA to infer the activity of TFs during discrete stages of differentiation, through the use of the Molecular Signatures Database (MSigDB) compendium of TF target gene sets (39). The MSigDB c3 TFT collection consists of 615 gene sets, and many transcription factors are represented by multiple gene sets (18). We used stringent criteria to define TFs with enriched activity, requiring more than 50% of corresponding target gene sets to be significantly enriched (FDR < 0.25) and more than one to be highly significant (FDR < 0.01).
Evaluation of Yy1 Binding.
Publicly available ChIP sequencing (ChIP-seq) data from human cells was used for preliminary validation of the murine GCB-specific Yy1 target genes highlighted by HG-GSEA. Raw Yy1 ChIP-seq signal data from K562 cells were obtained (30) using the UCSC table browser (40) for regions 2 kb upstream and 2 kb downstream of predicted Yy1 target genes. The presence of Yy1 binding peaks was determined as previously described (30). A subset of the predicted Yy1 target genes was further validated using chromatin ChIP-coupled quantitative real-time PCR (ChIP-qPCR) using ChIP assay kit (Upstate Biotechnologies) and available quantitative PCR assays (Applied Biosystems). Detailed protocols are described in SI Materials and Methods.
DNA Sequence Motif Analysis.
We conducted a comprehensive analysis of the enrichment of Yy1 binding sequence motifs in GCB signature genes and representation of this motif in all signature genes. This was performed by obtaining promoter region sequences, 2 kb upstream/downstream of the transcription start site, for each of the GCB signature genes using the UCSC table browser (40). Promoter regions were analyzed for significantly overrepresented DNA sequence motifs (FDR < 0.1), and motifs with ≥75% identity were combined into clusters using CisFinder (31). Clusters were then correlated with known TF binding matrices from the CisView and TransFac databases to identify the regulatory transcription factor (31, 41). The regulatory regions of GCB signature genes and the combination of all other signature genes were then searched for occurrence of the identified Yy1 consensus binding motif using CisFinder. The frequency of occurrence of the Yy1 motif by physical position within GCB and non-GCB signature genes was derived using CisFinder.
Evaluation of Murine GCB-Specific Yy1 Target Genes in Human GCBs.
Specificity of murine GCB Yy1 target genes for human GCB cells was evaluated using a publicly available dataset consisting of human naive B cells, centroblasts, centrocytes, and memory B cells (GEO accession no. GSE2350) (42). Human GCB (centrocytes and centroblasts) cells were compared with non-GCB (naive and memory) cells for the expression of murine GCB-specific Yy1 target genes, defined by DNA sequence motif, using gene set enrichment analysis with GSEA-P software (39).
Analysis of B-Cell Differentiation Signatures in Murine B-Cell Lymphoma Models.
We evaluated B-cell differentiation signatures in three murine B-cell lymphoma models that share common features with Pro/Pre-B (Lig4/p53 knock-out) (32), GCB (BCL6 knock-in) (33), and post-GCB (Bcl6/Myc-transgenic) (34) cells using Affymetrix Mouse 430A 2.0 microarrays. Detailed descriptions of sample processing are provided in SI Materials and Methods. Transcriptional signatures for each tumor model were derived using a one-versus-all analysis compared with the remaining two models, as described above, and the top 350-markers according to FDR Q-value.
Classification signatures of the nonmalignant B-cell subsets were derived by adding a fold-change criteria (FC > 1.25) to identify the most robust markers within the more comprehensive 350-gene signatures. Because of the similarities in gene expression profiles between Pro-B and Pre-B subsets and between Follicular B and MZ B subsets, these categories were combined into single classification signatures (Pre/Pro-B, Foll./MZ). The representation of normal B-cell classification signatures within tumor model signatures was assessed using HG-GSEA, as described above.
Acknowledgments
We thank Drs. Carlo Croce, Fritz Melchers, Max Cooper, and Barry Sleckman for critical review of this manuscript. This work was supported by National Institutes of Health Grant CA92625. F.W.A. and T.G. are Investigators of The Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper has been deposited in the GeoArchive database (GEO accession nos. GSE2350 and GSE26408).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1019537108/-/DCSupplemental.
References
- 1.Jung D, Giallourakis C, Mostoslavsky R, Alt FW. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu Rev Immunol. 2006;24:541–570. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- 2.Chaudhuri J, et al. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv Immunol. 2007;94:157–214. doi: 10.1016/S0065-2776(06)94006-1. [DOI] [PubMed] [Google Scholar]
- 3.Allman D, Pillai S. Peripheral B cell subsets. Curr Opin Immunol. 2008;20:149–157. doi: 10.1016/j.coi.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hagman J, Lukin K. Transcription factors drive B cell development. Curr Opin Immunol. 2006;18:127–134. doi: 10.1016/j.coi.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 5.Klein U, Dalla-Favera R. Germinal centres: Role in B cell physiology and malignancy. Nat Rev Immunol. 2008;8:22–33. doi: 10.1038/nri2217. [DOI] [PubMed] [Google Scholar]
- 6.Heng TS, Painter MW. Immunological Genome Project Consortium The Immunological Genome Project: Networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 7.Mandel EM, Grosschedl R. Transcription control of early B cell differentiation. Curr Opin Immunol. 2010;22:161–167. doi: 10.1016/j.coi.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 8.Lin YC, et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol. 2010;11:635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alizadeh AA, et al. Distinct types of diffuse large B cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 10.Kim TM, Chung YJ, Rhyu MG, Jung MH. Inferring biological functions and associated transcriptional regulators using gene set expression coherence analysis. BMC Bioinformatics. 2007 doi: 10.1186/1471-2105-8-453. 10.1186/1471-2105-8-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hardy RR, Kincade PW, Dorshkind K. The protean nature of cells in the B lymphocyte lineage. Immunity. 2007;26:703–714. doi: 10.1016/j.immuni.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 12.Nutt SL, Kee BL. The transcriptional regulation of B cell lineage commitment. Immunity. 2007;26:715–725. doi: 10.1016/j.immuni.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 13.Schmidlin H, Diehl SA, Blom B. New insights into the regulation of human B cell differentiation. Trends Immunol. 2009;30:277–285. doi: 10.1016/j.it.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vikstrom I, et al. Mcl-1 is essential for germinal center formation and B cell memory. Science. 2010;330:1095–1099. doi: 10.1126/science.1191793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bracken AP, Helin K. Polycomb group proteins: Navigators of lineage pathways led astray in cancer. Nat Rev Cancer. 2009;9:773–784. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- 16.Maiuri MC, Criollo A, Kroemer G. Crosstalk between apoptosis and autophagy within the Beclin 1 interactome. EMBO J. 2010;29:515–516. doi: 10.1038/emboj.2009.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Latchman DS. Eukaryotic transcription factors. Biochem J. 1990;270:281–289. doi: 10.1042/bj2700281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie X, et al. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grossmann M, et al. The anti-apoptotic activities of Rel and RelA required during B cell maturation involve the regulation of Bcl-2 expression. EMBO J. 2000;19:6351–6360. doi: 10.1093/emboj/19.23.6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franzoso G, et al. Requirement for NF-kappaB in osteoclast and B cell development. Genes Dev. 1997;11:3482–3496. doi: 10.1101/gad.11.24.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson BE, Mochon E, Boxer LM. Induction of bcl-2 expression by phosphorylated CREB proteins during B cell activation and rescue from apoptosis. Mol Cell Biol. 1996;16:5546–5556. doi: 10.1128/mcb.16.10.5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knox KA, Johnson GD, Gordon J. Distribution of cAMP in secondary follicles and its expression in B cell apoptosis and CD40-mediated survival. Int Immunol. 1993;5:1085–1091. doi: 10.1093/intimm/5.9.1085. [DOI] [PubMed] [Google Scholar]
- 23.Srinivasan L, et al. PI3 kinase signals BCR-dependent mature B cell survival. Cell. 2009;139:573–586. doi: 10.1016/j.cell.2009.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seto E, Shi Y, Shenk T. YY1 is an initiator sequence-binding protein that directs and activates transcription in vitro. Nature. 1991;354:241–245. doi: 10.1038/354241a0. [DOI] [PubMed] [Google Scholar]
- 25.Liu H, et al. Yin Yang 1 is a critical regulator of B cell development. Genes Dev. 2007;21:1179–1189. doi: 10.1101/gad.1529307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Srinivasan L, Atchison ML. YY1 DNA binding and PcG recruitment requires CtBP. Genes Dev. 2004;18:2596–2601. doi: 10.1101/gad.1228204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Galen JC, et al. Distinct expression patterns of polycomb oncoproteins and their binding partners during the germinal center reaction. Eur J Immunol. 2004;34:1870–1881. doi: 10.1002/eji.200424985. [DOI] [PubMed] [Google Scholar]
- 28.Schlisio S, Halperin T, Vidal M, Nevins JR. Interaction of YY1 with E2Fs, mediated by RYBP, provides a mechanism for specificity of E2F function. EMBO J. 2002;21:5775–5786. doi: 10.1093/emboj/cdf577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Q, Gedrich RW, Engel DA. Transcriptional repression of the c-fos gene by YY1 is mediated by a direct interaction with ATF/CREB. J Virol. 1995;69:4323–4330. doi: 10.1128/jvi.69.7.4323-4330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Euskirchen GM, et al. Mapping of transcription factor binding regions in mammalian cells by ChIP: Comparison of array- and sequencing-based technologies. Genome Res. 2007;17:898–909. doi: 10.1101/gr.5583007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharov AA, Ko MS. Exhaustive search for over-represented DNA sequence motifs with CisFinder. DNA Res. 2009;16:261–273. doi: 10.1093/dnares/dsp014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frank KM, et al. DNA ligase IV deficiency in mice leads to defective neurogenesis and embryonic lethality via the p53 pathway. Mol Cell. 2000;5:993–1002. doi: 10.1016/s1097-2765(00)80264-6. [DOI] [PubMed] [Google Scholar]
- 33.Cattoretti G, et al. Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B cell lymphomas in mice. Cancer Cell. 2005;7:445–455. doi: 10.1016/j.ccr.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 34.Pasqualucci L, et al. AID is required for germinal center-derived lymphomagenesis. Nat Genet. 2008;40:108–112. doi: 10.1038/ng.2007.35. [DOI] [PubMed] [Google Scholar]
- 35.Irizarry RA, et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003 doi: 10.1093/nar/gng015. 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Savage KJ, et al. The molecular signature of mediastinal large B cell lymphoma differs from that of other diffuse large B cell lymphomas and shares features with classical Hodgkin lymphoma. Blood. 2003;102:3871–3879. doi: 10.1182/blood-2003-06-1841. [DOI] [PubMed] [Google Scholar]
- 37.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fleiss J. Statistical Methods for Rates and Proportions. New York: Wiley; 1981. [Google Scholar]
- 39.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karolchik D, et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32(Database issue):D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wingender E, et al. TRANSFAC: An integrated system for gene expression regulation. Nucleic Acids Res. 2000;28:316–319. doi: 10.1093/nar/28.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Basso K, et al. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37:382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]