Figure 4.
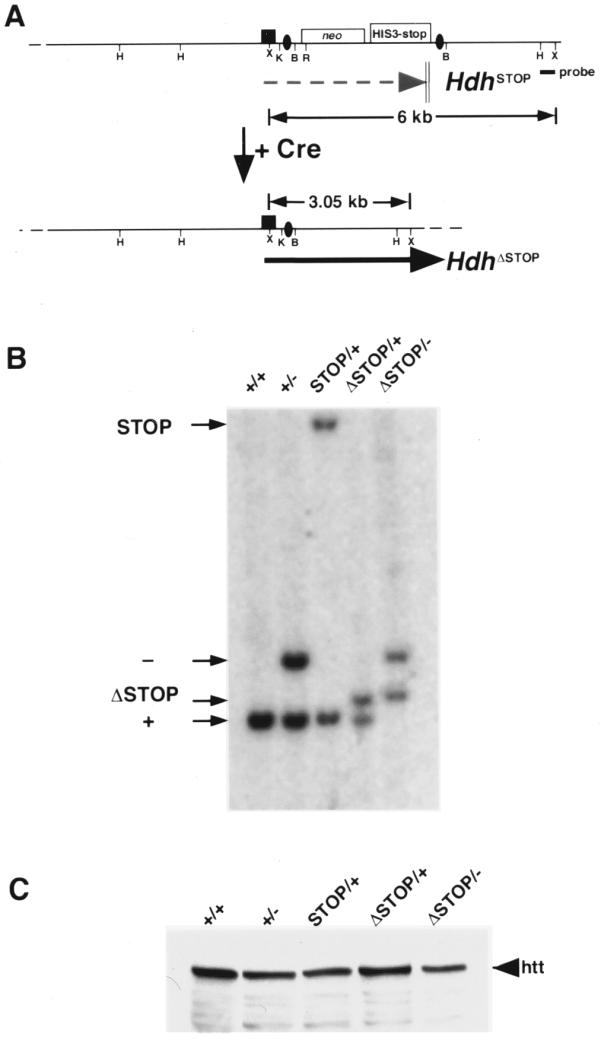
Cre-mediated excision of the neostop cassette restores Hdh expression. (A) Schematic of the HdhSTOP allele before and after (HdhΔSTOP) Cre-mediated recombination. Termination of the HdhSTOP transcript within the neostop cassette is indicated with a dashed gray arrow. Following Cre-mediated recombination, the neostop cassette is excised leaving a single loxP site (black oval) within the HdhSTOP intron 1 that does not interfere with expression [black arrow, see (C) below]. The position of a 3′ flanking probe that is able to distinguish between the HdhSTOP and HdhΔSTOP alleles is indicated with a black rectangle along with the sizes of diagnostic XmnI restriction fragments. Restriction sites are: HindIII (H), XmnI (X), KpnI (K), BamHI (B), EcoRI (R). (B) Southern analysis of tail biopsy DNA from wild-type (+/+), Hdh+/– (+/–), HdhSTOP/+ (STOP/+), HdhΔSTOP/+ (ΔSTOP/+) and HdhΔSTOP/– (ΔSTOP/–) mice. DNA digested with XmnI was blotted and hybridized with the 3′ flanking Hdh probe that recognizes the 6 kb STOP, 3.5 kb null (see Fig. 1C for a map of the Hdh– allele and diagnostic restriction fragments), 3.05 kb ΔSTOP and 3.0 kb wild-type genomic fragments. HdhΔSTOP/– mice were obtained in the predicted Mendelian ratio and were indistinguishable from their wild-type littermates. (C) Western analysis of cytoplasmic extracts prepared from brain tissue dissected from mice with the same genotypes analyzed in (B) using an anti-huntingtin antibody. The amount of huntingtin (htt) in brain extracts prepared from HdhΔSTOP/– mice was equivalent to the amount of huntingtin in Hdh+/– or HdhSTOP/+ extracts.
