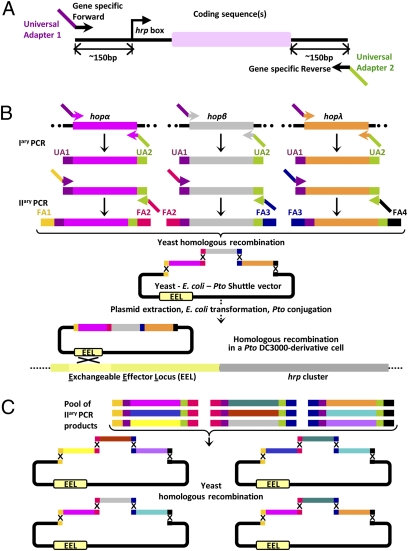
Fig. 4.
The programmable or random in vivo assembly shuttle (PRIVAS) system exploits dual adapter recombination for facile integration of combinatorial gene sets into the DC3000 exchangeable effector locus (EEL). (A) Structure of a typical T3E genetic unit (GU) for PRIVAS. Primary PCR reactions with gene-specific oligonucleotide primers harboring 20-bp 3′ extensions amplify GUs flanked on each side by universal adaptor (UA) regions 1 or 2. (B) Secondary PCR reactions use these primary products as templates and flexible adapter (FA) primers composed of UA-specific segments at their 3′ end and one of a set of ~35-bp FA-homology regions at their 5′ end to yield UA-FA dual adapter-flanked GUs that are used as the elementary building blocks for in vivo assembly in yeast. The configuration of the gene sets, including gene orientation, can be fully programmed during construction by designing FA-flanked GUs so that a unique combination of recombination events between FAs leads to the closure of a circular DNA molecule containing the sequences of the shuttle vector as depicted in B for 3 GUs. The shuttle vector's backbone provides the origins of replication and selection markers for yeast and E. coli as well as an origin of transfer for conjugation into P. syringae. After transformation with a suitable pool of GUs and the linearized shuttle vector, plasmid DNA is extracted from yeast cells surviving selection and transferred into E. coli for subsequent conjugation into a recipient P. syringae strain and single cross-over integration into the EEL. (C) PRIVAS also can be used in random mode for the creation of complex combinatorial libraries of gene sets of variable configuration but of fixed size (equal to three in C). If several distinct GUs sharing the same pair of external FAs specifying a given position within the gene sets are included in the assembly reaction, identical FAs compete for recombination and hence, incorporation in growing DNA molecules. The final circular products contain polymorphic sets composed of GUs drawn from distinct bins of GUs at desired positions as illustrated in C.