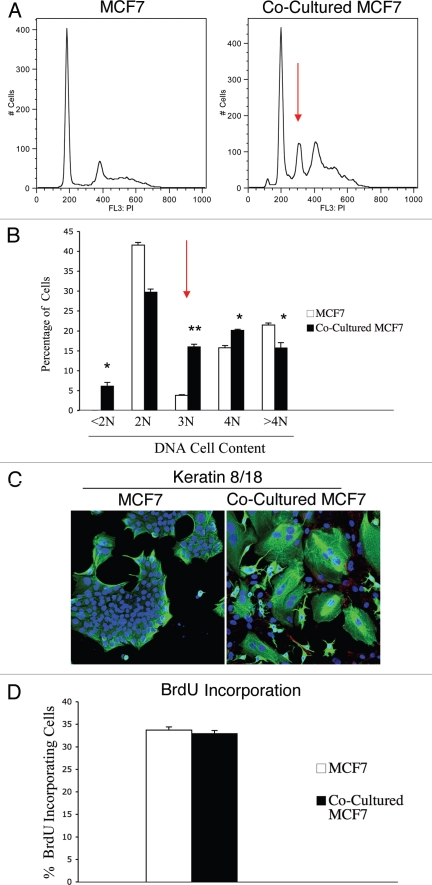
Figure 5.
Fibroblasts induce aneuploidy in co-cultured MCF7 cells. (A and B) DNA cell content analysis. GFp (+) MCF7 cells were plated in mono-culture or in co-culture with hteRt-fibroblasts. the day after, media was changed to DMEM with 10% NuSerum and cells were grown for 48 hours. then, to isolate the GFP (+) MCF7 cell population from the fibroblasts, co-cultured cells were FACS-sorted using a 488 nm laser. As a critical control, mono-cultures of GFP (+) MCF7 cells were sorted in parallel. Then, sorted cells were fixed and stained with PI. DNA cell content was analyzed by flow cytometry. (A) Representative traces. Note that co-culture with fibroblasts induces the appearance of a large aneuploid peak (cells with an abnormal DNA content of ∼3N) in MCF7 cells (red arrow). In addition, a larger population of cells with DNA content of 4N is detected in co-cultured MCF7 cells compared to MCF7 cell mono-cultures. (B) Cell numbers with a given DNA cell content were determined and graphed as a percentage of the total population. *p < 0.01, **p < 0.0000003. (C) Immunofluorescence. MCF7 cells were grown in mono-culture or in co-culture with hTERT-fibroblasts for 5 days. Then, cells were fixed and immunostained with antibodies directed against Cav-1 (red, labeling fibroblasts) and K8/18 (green, labeling MCF7 cells). Nuclei were counterstained with DAPI (blue). Note that many MCF7 cells in co-culture are multinucleated, suggesting that in co-culture a sub-population of MCF7 cells undergo nuclear, but not cellular division. Note also that Cav-1 expression is very low in co-cultured fibroblasts, as expected. original magnification, 20x. (D) BrdU incorporation. To evaluate proliferation, GFP (+) MCF7 cells were grown in co-culture with hTERT-fibroblasts or in mono-culture for 48 hours. Then, cells were pulsed-labeled with BrdU for one hour, sorted to purify the GFP (+) cells and fixed overnight. BrdU incorporation was analyzed by flow cytometry. Co-culture with fibroblasts does not affect proliferation of MCF7 cells. Columns, relative BrdU incorporation from at least three independent experiments; bars, SEM.