Abstract
Many methods and tools have evolved for microarray analysis such as single probe evaluation, promoter module modeling and pathway analysis. Little is known, however, about optimizing this flow of analysis for the flexible reasoning biomedical researchers need for hypothesizing about disease mechanisms. In developing and implementing a workflow, we found that workflows are not complete or valuable unless automation is well-integrated with human intelligence. We present our workflow for the translational problem of classifying new sub-types of renal diseases. Using our workflow as an example, we explain opportunities and limitations in achieving this necessary integration and propose approaches to guide such integration for the next great frontier-facilitating exploratory analysis of candidate genes.
Introduction
In the Kretzler laboratory, one main focus is to discover through a systems approach previously unknown renal sub-diseases. These discoveries can significantly advance clinical diagnosis, screening, and therapies. Our flow of research involves numerous analytical processes and applications and is conducted over many months in conjunction with local and remote collaborators. Because of the diverse data resources and tools required end-to-end and because of the great amount of data sharing that goes on, numerous errors from manual computations and exchanges are possible. Thus we have studied our research processes closely and have developed and implemented automated workflows for greater quality assurance and efficiency.
Much research on scientific and systems biology workflows shows that creating workflow modules for recurring, time-consuming, formalizable tasks and computations is fairly straightforward technically. Yet many unmet challenges still exist for integrating these automated workflow modules with the non-formalizable tasks of scientific discovery. These challenges involve human issues and imply nontrivial technical solutions.
Our self-study offers insights into two of these unmet challenges.(a) Modeling and designing for the human-in-the-loop in formalizable modules, and (b) modeling the mixed formal and nonformal aspects of scientists’ exploratory analysis. Both models are needed to assure systems that match scientists’ knowledge representations and interaction needs. In regard to these challenges, our self-study confirms a growing but not yet dominant school of thought, namely that the requirements for adaptability and flexibility for the first challenge differ from those of the second challenge. In this article, we show the different kinds of adaptability and propose the research that is still needed for accomplishing especially the second – integrating support for structured and nonstructured analytical tasks.
Related Work
Technically, scientific workflows are “processes that combine data and computational processes into a configurable structured set of steps for automated solutions to a scientific problem” [13]. Developers typically decompose scientists’ analytical tasks into standardizable components, build these components into a set of procedures, information flows, and tools; and have humans or automated programs carry out this set of processes [2]. However, for scientific discovery and hypothesizing, “human and organizational aspects … are equally critical for success as technical issues” and require building adaptability into workflows [2].
Much of the adaptability required for discovery-based workflows is outside the scope of our self-study, for example provenance issues [3]. But within our scope are two needs. The first is to adaptively keep the human in-the-loop in managing information and sensemaking, e.g. letting users decide parameters and the usage of certain resources, setting boundaries for decisions a system can make without user involvement, and letting users identify and select data and context to carry over for collaborations [4–5]. Research shows that this adaptability involves building in awareness of domain-specific choice points, a non-trivial problem [6].
A second aspect of adaptability relevant to our case is assuring that routine processes in analysis are integrated with the partially or underspecified flow that invariably occur in explanatory analysis [6]. Researchers note that this adaptability for scientific discovery is only in its earliest stages, both in terms of user modeling and system design [7]. For the routine and formalizable parts of scientific discovery in bioinformatics, some user models have been developed, for example, those that capture bioinformatics specialists’ protocols for conceptually differentiating relationships when categorizing gene sequence data [8–9]. Non-formalizable episodes are harder to model. They involve the contextually-driven dimensions of interpretations, inferences, analogies, validations, and opportunistic creative leaps. Dourish and Edwards argue that integrating users’ formal and nonformal dimensions of analysis requires “radical degrees of flexibility” [7,48]. Workflows have to be aware of and dynamically respond to changing circumstances and have to be able to operate over many layers of scientists’ work and domain knowledge.
To reach these ends, technological strategies and techniques range from Stevens et al’s patterns for supporting queries to diverse sources to sophisticated semantic indexing and data mining by concept and wrappings [10–12]. The success of any of these solutions, however, ultimately depends on having and linking them to empirically sound models of scientists’ explorations and explanations [12]
Overview of the Case Study Workflow
To better understand and reveal current strengths and gaps in electronic workflows while concurrently gaining efficiencies and error reductions that workflows offer, we developed and implemented workflow modules (called processes here). These relate analyzing patient histological data and expression data at a systems level to gain novel insights about courses of different types of renal disease and to hypothesize about their underlying mechanisms.
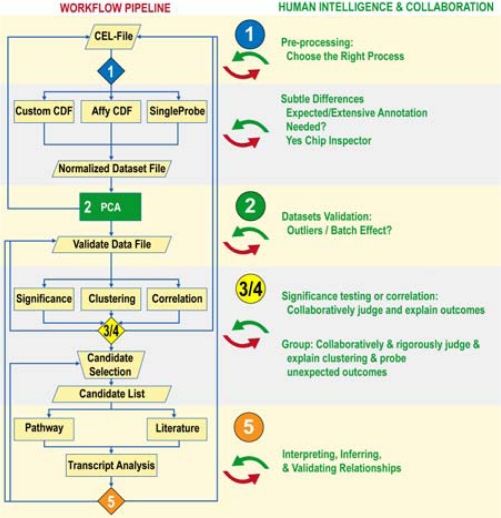
Five analytical processes comprise this workflow, as detailed below and in Figure 1. The first four lend themselves to automation and uses of GenePattern (http://www.broad.mit.edu/cancer/software/genepattern/). The fifth process – explanatory analysis under uncertainty for hypothesizing purposes – requires continuous human interaction with dynamic data relationships with conceptual traits to find a credible discovery path amid biological contexts and constraints. For this process, we have used various tools to semantically mine the research literature and to infer biological events through analyses ofpathways, promoters, and protein interactions. As we show, for this process, workflows that connect inputs and outputs from various analysis applications are still a long way from productively integrating human complex reasoning and automation.
Figure 1.
Workflow diagram
Processes 1–4: Mutually Related Automation and Human Intelligence
1. Data pre-processing involves normalizing datasets from different arrays. First, specialists raise and answer several questions that cannot be automated, such as:
- What is the most comprehensive set of CEL files that we expect to be comparable?
- Are there quality problems with CEL file(s) that warrant excluding it?
- Do we expect the analysis to go beyond the capabilities of our standard procedure (analysis on the gene level) and thus require us to change to methods with a higher resolution (single probe evaluation /transcript level, ChipInspector)
Then files get normalized through different automation methods; depending on size or the need for high resolution. In some methods – e.g. those using GenePattern - normalization is transparent, thus facilitating the evaluation of output for accuracy and completeness. Methods with extensive annotation, such as ChipInspector, also facilitate evaluations but with trade-offs: As proprietary software, ChipInspector is neither entirely transparent nor state of the art in normalization.
Finally, normalized datasets/output get validated. Earlier choices about normalization methods affect specialists’ interpretations. Specialists also must fit into the loop to determine how to store the data in proprietary sources and manage/coordinate file versions for accuracy.
2. In validating the integrity of datasets, Principal Component Analysis (PCA) is run, and specialists again must be in the loop to critically examine the graphic and tabular output. They define clusters and sub-groupings and determine causes of problems such as outliers. They ask, e.g: Are the data degraded - is it a matter of removing specific CEL files and rerunning the preprocessing? Or if the data are valid, might some other feature of the data – such as an animal dataset being included when it should not be - explain outliers? Alternately, might a batch effect be the problem? If so, intensive analysis among collaborators ensues, often from many disciplines.
Several iterations of pre-processing→PCA may be needed. For us, automating portions of this iteration expedited what had previously been an extremely lengthy process. Before we automated these processes, we had to use different programs with different data formats. Doing so introduced unnecessary data conversions, a process that cost time and increased the risk of errors.
In validating datasets, human expertise cannot be automated for such nonformalizable reasoning as scrutinizing output and finding, diagnosing and resolving problems.
3. Grouping aims to identify “gene expression fingerprints” associated with patients. Samples are grouped through automation by two different clustering approaches. The first includes but is not limited to unsupervised hierarchical clustering and evaluates if structures found in molecular data can be mapped to patient information such as histological data. A special point of interest is to search for molecule-distinct subcategories within a histological classification, which could indicate different courses of a disease or different responses to treatment.
The second method is entirely data-based and seeks to find a classification scheme for patients only on their molecular data, an approach that could either confirm or challenge the current disease classification.
In either case, human expertise is needed to determine the appropriate clustering method and parameters and to evaluate the implications of the methods on the results. Additionally, humans’ domain knowledge is needed to set the results into a disease context. For these analyses downloadable output is crucial. The ways in which output is represented can bias interpretations; hence scientists need to validate their interpretations against more than one representation.
4. Significance testing or correlation uses automation to filter out potentially informative genes for further study. Independently of groups, we use correlation and ridge regression as methods to connect genes and clinical parameters and to establish marker sets that can predict course of disease.
Contrarily in case of a given classification we use significance analysis to reveal the genes that are most distinct between predefined classes. This analysis shifts the research focus from prediction to functional differences.
As specialists examine the data and find problems they often need to bring in other people. For example, if several probesets representing a gene of interest are discordantly regulated microbiologists are consulted about alternative splicing, and iterations of normalization with higher resolution may be run.
Output of Process 4 is a prioritized candidate gene list, the initial seed for interactively mapping genes and gene products onto contexts that may explain biological events and courses of disease types.
Process 5: What Human Intelligence Corresponds to Automated Output?
The fifth process involves analyzing candidate genes within dynamic relationships and contexts to interpret, infer, and validate causal associations and hypotheses about mechanisms of sub-type diseases. As biology works in networks and modules, individual genes carry only limited information. This process thus relies on several tools to relate findings from prior steps and to situate analysis in biologically meaningful contexts.
Protein interaction applications such as MiMI (mimi.ncibi.org) can reveal and highlight literature about multidimensional relationships of interest based on correlated attributes. Other tools offer statistical assurances and localization.
Pathway analysis tools as Ingenuity (www.ingenuity.com) map genes to already established modules referred to as canonical pathways or use current knowledge from literature and experiments to find functional relations between them. While this provides a very broad context for the relations of the input genes, we typically lack crucial detailed information as intergenic relations in the specific tissue and the role of individual transcripts. Additionally, the use of current knowledge with its unequal distribution can bias the results, a problem that holds true for literature mining as for experiment databases.
Alternately, networks can be generated initially from literature analysis (Genomatix BiblioSphere, http://www.genomatix.de/) for subsequent promoter analysis (Genomatix’s FrameWorker). Initial results are reduced and confirmed by close investigation of the DNA based on the spacing and order of structural elements. The output, promoter modules, can be mapped to the promoters of all genes and thus extend our functional knowledge. But a break in scale similarly occurs. Network generation and analysis are performed on a gene level; promoter modules are defined on a transcript level. This causes problems in both directions: The network on gene level can be too coarse to maintain enough information for the extraction of a meaningful promoter module. The information gained in transcript level cannot be mapped back to the network. This analysis requires support for users’ interpretations and inferences across scales on many dimensions, not just one level.
Human expertise and human rigor in prior processes are crucial for gene selection when, as with FrameWorker, input cannot exceed 10–20 genes due to computational limitations. Such restrictions mean that a flawless data exchange from prior data generation and interpretation must transpire. It is mandatory to find the optimal data representation, to limit interpretation to the data content, and to validate results.
Summary / Discussion of Integrated Processes
Our workflow case reveals that most of the steps from Processes 1–4 can be integrated into the GenePattern framework for transparency, flexibility and reproducibility. GenePattern provides apt standardization. The consistent file formats in GenePattern reduce data transformations to a minimum, eliminate error sources and the possible omission of data integrity checks greatly improves analysis speed from several days to hours, not counting the decision making. Its modular structure offers flexibility for coupling workflows with collaborative interpretations by experts across sub-specialties. Filling in a gap in the current research literature, our case study gives concrete form to questions and choice points at which human intelligence and collaboration must enter the picture.
Our study also helps to specify four traits that scientists’ analysis processes should have if they are to be decomposed and automated. They should: (a) involve computations that machines can do fast and reliably; (b) have no need for intervening human intelligence or choice during computations; (c) generate output that does not raise threats of bias during interpretation, either because automated methods are assumed statistically to be unbiased (e.g. Principal Component Analysis) or because different automated methods are built in and provide diverse enough perspectives on the data to gain a complete and credible picture; and (d) involve a set of well-defined questions that specialists commonly and reliably apply to computations.
However, we found that even processes or modules lend themselves to automation require mechanisms for adaptively including human expertise. For the first four processes in our workflow, for example, a rigorous quality check after each step enables domain specialists to minimize errors. It allows researchers to get familiar with the attributes of the specific dataset, to optimize its treatment, and to ask the appropriate questions.
One reason why built-in adaptability for Processes 1–4 is necessary is that trade-offs associated to a set of automated methods are often revealed only several steps afterwards. For example, while normalization on gene level is sufficient for most purposes, it can lead to a situation in which several probesets representing one gene show discordant correlation/regulation. Normalization on transcript or even exon level can help to address these issues by increasing the resolution but it lowers coverage and confidence since the transcripts/exons are represented by fewer probes. As a result, combined automation and human intelligence for these analyses can be a highly iterative process, with efficiencies elucidating facets of the dataset more quickly, thus giving scientists more time for creative exploration.
The fifth process - interpreting, inferring and validating candidate genes – is in many ways a different story. In this process, biomedical researchers strive to uncover explanatory associations within and across levels of biological systems, and dynamic causal events in these systems are complex, often uncharted and ambiguous. It is uncertain which components can be separated from human exploratory analysis or what output reduces threats of bias. For example, topological statistics can be run on protein-protein interaction networks (e.g. cluster densities, recurrent motifs) but biomedical researchers are hesitant to go further without knowing (a) whether the configurations are by chance alone and (b) what these statistics imply in regard to biological meaning. These are open questions. We do not know yet what workflows must offer in order to assure specialists that they have the optimal representations, workspaces and analytical interactions for negotiating inputs, outputs, and biases effectively. For this fifth process, how to exploit automation and integrate it with human reasoning is a nontrivial challenge.
Conclusion
Connecting standard processes to local pipelines and collaborative interpretations by domain experts has helped us achieve efficiency, quality, credibility, and discovery. Based on the human and automated processes vital to the workflow described here, several insights have emerged from this project about requirements for adaptability for effectively keeping the human-in-the-loop . They include
Any data produced by workflow processes must be downloadable for the team to analyze further
Any workflow tool must support on-demand analysis by researchers across sub-specialties.
On-demand analysis and resolutions depend on the level at which a problem is found.
Integration requires open channels of communication, actual/virtual social proximity and regular exchanges among molecular biologists, bioinformaticians, biostatisticians, biomedical researchers, clinical researchers, PIs, database experts, and tool experts
Pipelines have significantly reduced the pure data processing and implemented techniques to help the researcher find the appropriate question but Process 5 reveals the next great frontier for workflow modeling and design with appropriate adaptability. While an application may provide a basic context through pathways and promoter modules, existing tools often fail to shape out the structure inherent in the specific dataset clearly enough to be used as base for hypothesis generation.
Our study shows that better models of scientists’ actual complex analysis processes are needed and that better data mining of concepts and relationships . Also our study suggests that choices of grain size for naming processes go hand-in-hand with designs for coordinating and integrating modules and that the disparate outputs and inputs relevant to complex tasks must be specified in ways that assure analytical coherence.
Acknowledgments
The study was supported by the following grants to MK: NIH # P30-DK081943-01 and UM CTSA multidisciplinary pilot grant. It also was supported by NIH Grant #U54DA021519. We thank Sebastian Martini and Christian Albiker for their assistance
References
- 1.Altinas I.New challenges for user-oriented scientific workflowsIn NSF Workshop on challenges of ScientificWorkflows. 2006. Last accessed on 1/28/09: http://vtcpc.isi.edu/wiki/images/f/fe/ParticipantStatements.pdf:
- 2.Merrill J. Listserv entry to AMIA’s poi-wg maillist. Dec 21, 2006.
- 3.Anderson NR, Lee ES, Brockenbrough JS, Mimie M, Fuller S, Brinkjley J, Tarczy-Hornoch P. Issues in biomedical research data management and analysis: needs and barriers. JAMIA. 2007;14:478–488. doi: 10.1197/jamia.M2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scacchi W.Discovering, modeling, analyzing and re-enacting scientific work processes and practicesIn NSF Workshop on Challenges of Scientific Workflows 2006; Last accessed on January 28, 2009: http://vtcpc.isi.edu/wiki/images/f/fe/ParticipantStatements.pdf
- 5.Jorgensen HD. Interaction as a framework for flexible workflow modeling. Proceedings of ACM GROUP ’01. 2001:32–41. [Google Scholar]
- 6.Divitini M, Simone C.Supporting different dimensions of adaptability in workflow modeling CSCW2009365–397. [Google Scholar]
- 7.Dourish P, Edwards WK. A tale of two toolkits: relating infrastructure and use in flexible CSCW toolkits. CSCW. 2000;9:31–51. [Google Scholar]
- 8.Bartlett JC, Toms EG. Developing a protocol for bioinformatics analysis: an integrated information behavior and task analysis approach. JASIST. 2005;56:469–482. [Google Scholar]
- 9.Tran D, Dubay C, Gorman P, Hersh W.Applying task analysis to describe and facilitate bioinformatics tasks Proceedings of MEDINFO 2004 2004107(Pt2):818–822. [PubMed] [Google Scholar]
- 10.Gil Y, Deelman E, Ellisman M, Fahringer T, Fox G, et al. Examining the challenges of scientific workflows. IEEE Computer. 2007;40:24–32. [Google Scholar]
- 11.Stevens R, Goble C, Baker P, Brass A. A classification of tasks in bioinformatics. Bioinformatics. 2001;17:180–188. doi: 10.1093/bioinformatics/17.2.180. [DOI] [PubMed] [Google Scholar]
- 12.Landauer C, Bellman K. Wrappings for one-of-a-kind software development. Proceedings of the 35th Hawaii International Conference on System Sciences; 2002. [Google Scholar]