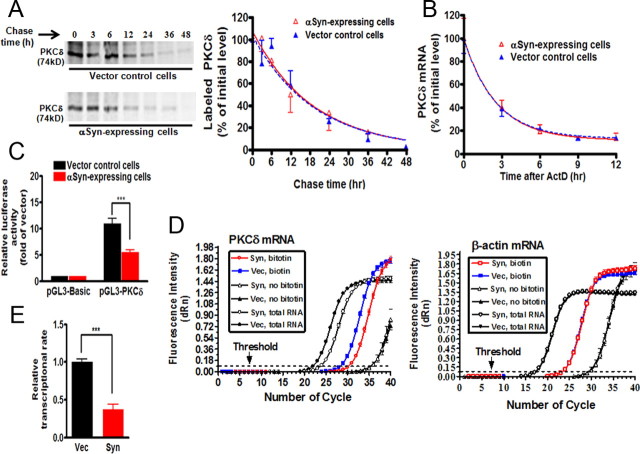
Figure 4.
α-Synuclein suppresses PKCδ transcription without affecting PKCδ protein or mRNA stability in N27 dopaminergic cells. A, Left, Pulse-chase analysis of stability of PKCδ protein. αSyn-expressing and vector control N27 cells were labeled with [35S]methionine, and PKCδ protein was analyzed over 48 h as described in Materials and Methods. Right, The bands were quantified and expressed as percentage of amount present at time 0 h. The data plotted were fit to a one-phase exponential decay model using the nonlinear regression analysis program of Prism 4.0 software as follows: Y = span e−Kt + plateau, where Y starts at span + plateau and decays with a rate constant K. The half-life of the protein was determined by 0.693/K. The square of the correlation coefficient (R2) is used as a measure of goodness-of-fit in regression analysis. Values are mean ± SEM of two independent experiments. B, The stability of PKCδ mRNA was not decreased in αsyn-expressing N27 cells. After treatment with ActD, total RNA was extracted for qRT-PCR analysis at selected time intervals. The relative abundance of PKCδ mRNA was expressed as a percentage of that present at time 0 h, and data plotted were fit to the one-phase exponential decay model. Values are mean ± SEM of three independent experiments performed in triplicate. C, The PKCδ promoter activation was attenuated in αsyn-expressing cells in reporter assays. Reporter pGL3-PKCδ carrying the PKCδ promoter or pGL3-Basic empty vector was transiently transfected into αsyn-expressing and vector control cells. Cells were collected 24 h after transfection and assayed for luciferase activity and β-galactosidase activity. Data were normalized and expressed as fold induction over the pGL3-Basic vector. Values are shown as mean ± SEM of three independent experiments performed in triplicate (***p < 0.001). D, The relative transcription efficiency of PKCδ was examined by quantitative nuclear run-on assay. Representative amplification plots for PKCδ mRNA (left panel) and β-actin mRNA (right panel) are shown. The change in fluorescence intensity (ΔRn) was plotted on the y-axis. The arrow shows the threshold (dashed lines). E, Quantitation of transcription efficiency. Data are expressed as fold change in the level of nascent run-on PKCδ mRNA in vector control cells and are shown as mean ± SEM of three independent experiments performed in triplicate (***p < 0.001).