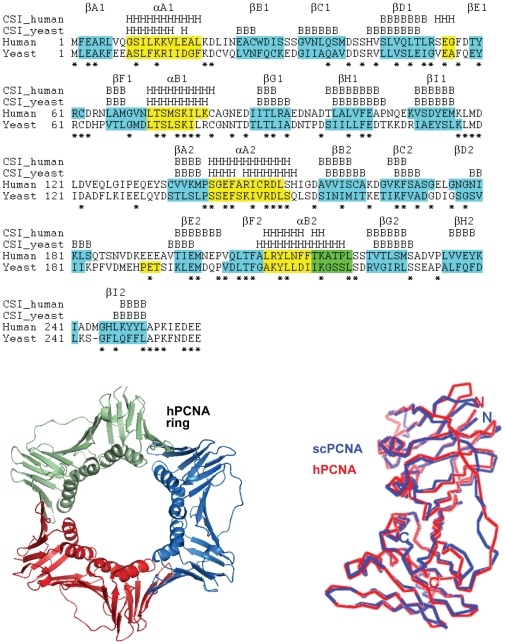
Figure 1. Structure of the PCNA sliding clamps.
Amino acid sequence alignment of human and yeast PCNA monomers. The conserved residues are indicated with an asterisk. The secondary structure elements within the two topologically identical domains were assigned from the crystal structures (PDB entries 1VYM and 1PLQ), sequentially labeled βA1, βB1, etc. (first domain) and βA2, βB2, etc. (second domain) and highlighted (light blue for β strands and yellow for α helices, in green are indicated residues annotated in a 3/10 helix conformation). The secondary structure of yeast and human PCNA molecules identified in solution using the Chemical Shift Index [44] obtained from the NMR chemical shifts is indicated above the sequences. H stands for α-helix and B for β-sheet. This index relies on the consensus of only three measurements at each position (only the 13Cα, 13Cβ, and 13C' were assigned, but not the 1Hα), and therefore there is a large number of residues for which an overall consensus could not be reached. At the bottom-left the superposition of the Cα trace of human PCNA (red) and yeast PCNA (blue) subunits is shown. At the bottom-right, a ribbon representation of the human PCNA trimeric ring is shown. Structures were prepared using PyMol (http://www.pymol.org).