Fig. 1.
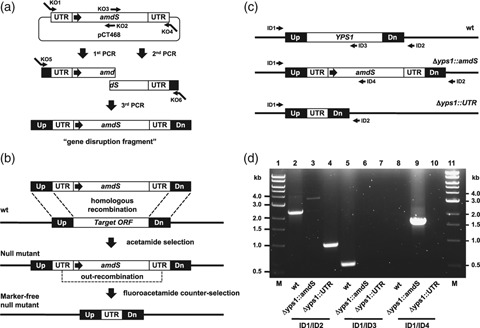
Gene deletion and amdS marker recycling strategy. (a) A linear gene disruption DNA fragment was assembled by three rounds of PCR. The fragment contained the Aspergillus nidulans amdS gene flanked by directly repeating 300-bp segments of its native 3′ untranslated region (UTR) and typically 160–250 bp of DNA homologous to regions upstream (Up) and downstream (Dn) of the target chromosomal locus. Expression of amdS was driven by the Saccharomyces cerevisiae ADH1 promoter (large horizontal arrow). (b) Upon its introduction into Kluyveromyces lactis cells, transplacement of the disruption fragment occurs at the target locus by homologous recombination resulting in gene deletion. Integrants are selected by growth on nitrogen-free medium containing acetamide. Subsequent out-recombination of the amdS marker occurs in the absence of selective pressure and amdS− null mutants are isolated by growth on counterselection medium containing fluoroacetamide. (c) PCR using a single forward primer (ID1) and various strategically positioned reverse primers (ID2-4) are used to assess the integrity of a modified locus. (d) An agarose gel showing an example of genomic PCR analysis of strains harboring Δyps1∷amdS and marker-free Δyps1∷UTR null alleles.
