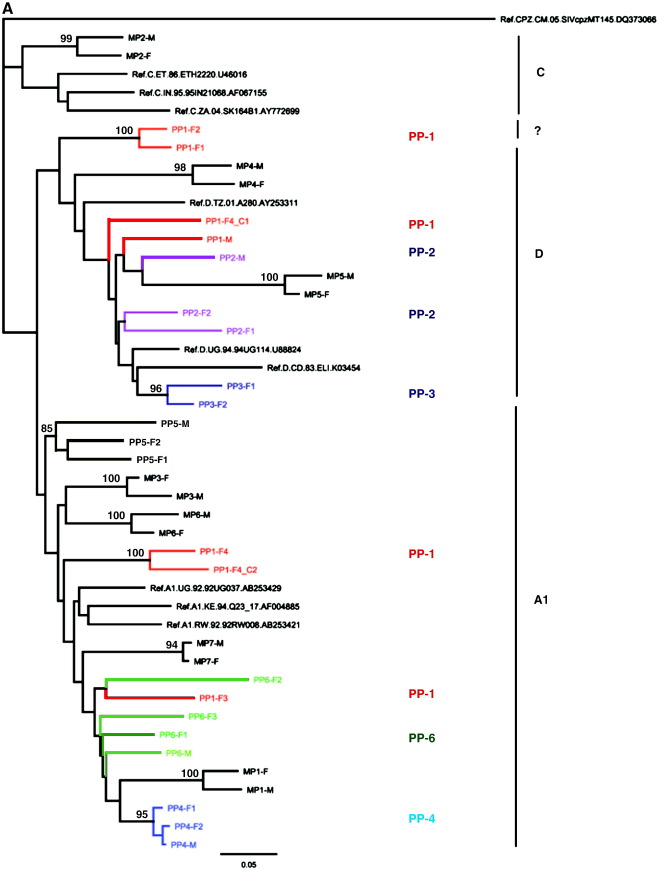
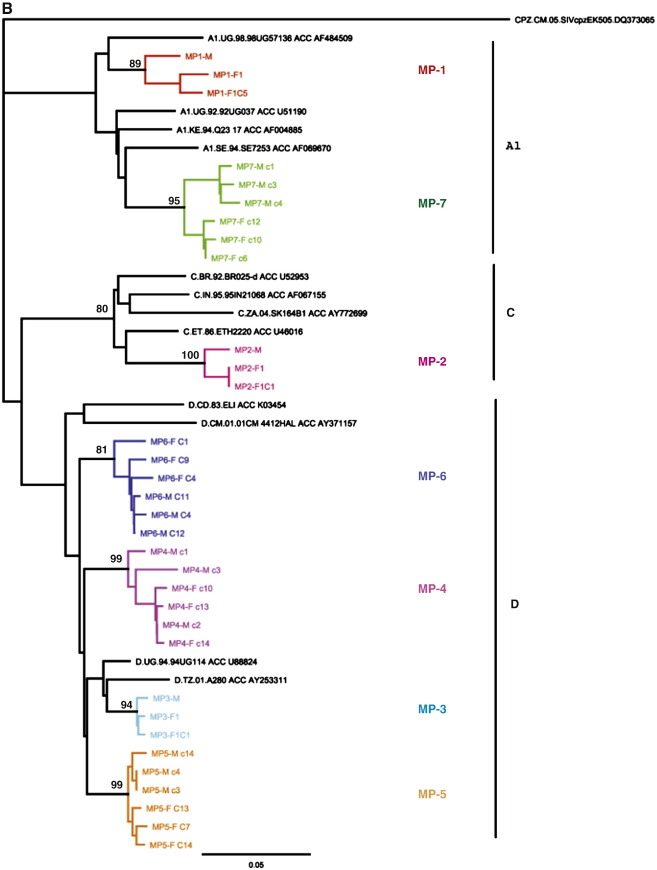
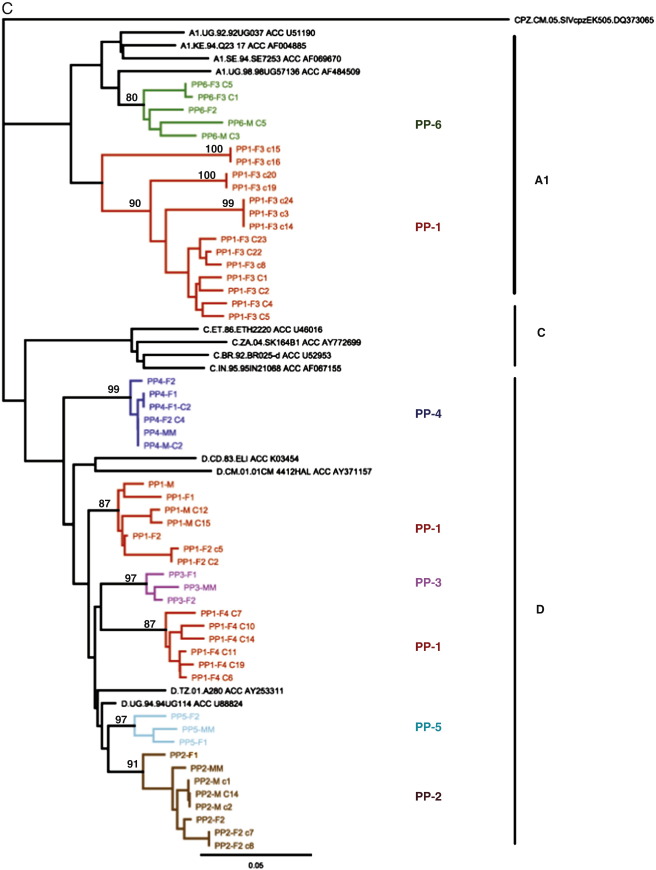
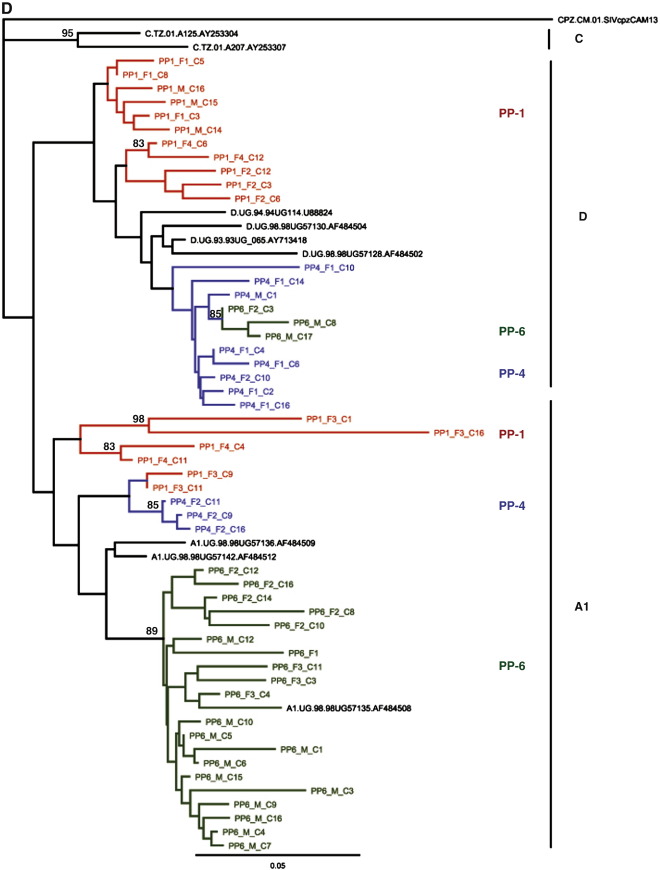
Fig. 1.
A. Phylogenetic analysis of the env-C2V3 sequences (HXB2 location 6829–7334) of Monogamous Partnerships (MP) 1–7 and Polygamous Partnerships (PP) 1–6. All PPs are coloured PP1 (red), PP2- (purple), PP3 (dark blue), PP4 (light blue), PP5 (black) and PP6 (green). Bootstrap values greater than 80% are shown. M—male partner and F—female partner. The phylogenetic plots do represent consensus sequences of the patients. B. Phylogenetic analysis of the gag-p24 sequences (HXB2 location 1123–1589) of Monogamous Partnerships (MP) 1–7. Bootstrap values greater than 80% are shown. M—male partner and F—female partner. The phylogenetic plots do represent consensus sequences of the patients. C. Phylogenetic analysis of the gag-p24 sequences (HXB2 location 1123–1589) of Polygamous Partnerships (PP) 1–6. Bootstrap values greater than 80% are shown. M—Male. D. Phylogenetic analysis of the pol-IN sequences in Polygamous Partnerships PP1, 4 and 6 (including clone and direct sequences HXB2 location 4470–4807). Bootstrap values greater than 80% are shown. M—male partner and F—female partner.