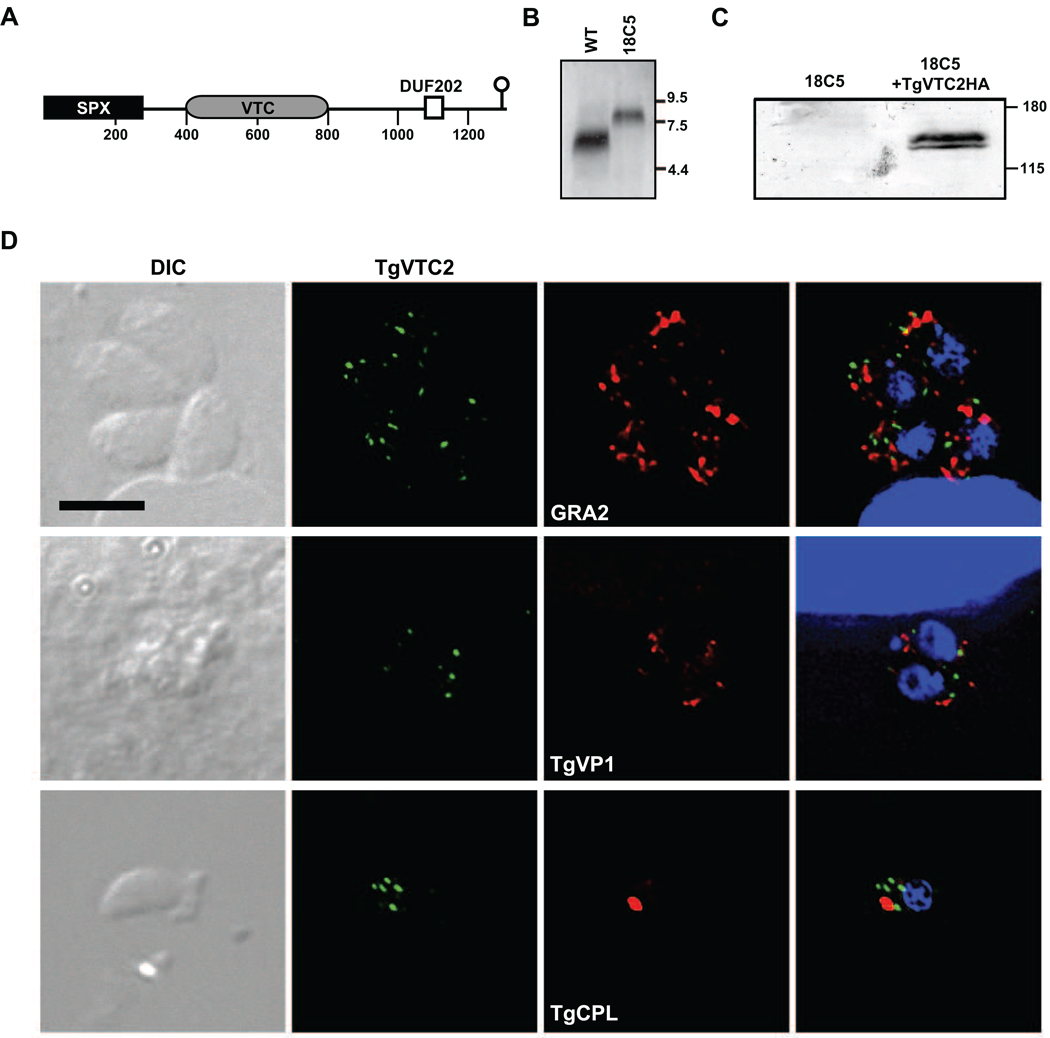
Figure 1.
A. Domain map of TgME49_098630 (TgVTC2). Shown are the relative positions of SPX, VTC and DUF202 domains. The site of insertion in strain 18C5 is indicated as a lollipop. Amino acid distance markers are shown under the cartoon.
B. Strain 18C5 is disrupted in predicted gene TgME49_098630. Parasites were grown in human foreskin fibroblasts (HFF) cultivated in Dulbecco’s modified eagle medium (DMEM) containing 10% fetal bovine serum (FBS). Total RNA was derived from approximately 1×107 intracellular tachyzoites of strains Pru (WT) and 18C5 by passage through a 27-gauge needle, centrifugation at 425×g for 10 min, and processing of the pellet with ULTRASPEC (BiotecX). RNA was separated by formaldehyde agarose gel electrophoresis, transferred to nytran, and probed with a radiolabeled (Random Primed DNA Labeling Kit, Roche) cloned DNA fragment amplified using primer 5’-CGATTCTCGTAGTCACACTGATGC and the 3’ RACE Inner Primer (FirstChoice RLM-RACE, Ambion Inc.). RNA size standards are indicated to the right in kilobases.
C. Amino-terminal HA-tagged TgVTC2 migrates as a dimer around 140 kDa. Of strains 18C5 and 18C5/pTPR2HA (18C5+TgVTC2HA), 2×107 extracellular tachyzoites lysed from HFFs were harvested by centrifugation at 425×g for 10 min, and pellets were suspended in denaturing protein sample buffer and boiled for 10 minutes. Samples were separated on 8% polyacrylamide gels, transferred to nitrocellulose, reacted with monoclonal anti-HA antibody (clone 16B12, #MMS-101R, Covance, Emoryville, CA), and detected with ECL Plus western blotting detection system (GE). Molecular mass standards are shown at the right in kDa.
D. TgVTC2 localizes within the cytoplasm. Confluent coverslips of HFFs were infected with tachyzoites of strain 18C5/pTPR2HA. Twenty-eight hours post-infection, monolayers were fixed with PBS/3% methanol-free fomaldehyde, blocked with PBS/3% BSA/0.2% triton X-100. TgVTC2 was detected with mouse monoclonal anti-HA (Covance) and AlexaFluor488. Subcellular compartments were defined with the following rabbit antisera: anti-MIC2 (micronemes), anti-GRA2 or anti-GRA7 (dense granules), and anti-TgVP1 (acidocalcisomes). Naturally egressed tachyzoites were fixed to coverslips and similarly reacted with rabbit anti-TgCPL (plant-like vacuole). These markers were detected with AlexaFluor633. Coverslips were mounted with VectaShield with DAPI (Vector Laboratories, Inc). Fluorescent images were captured using a Zeiss Axioplan 2 microscope and OpenLab 5.0.1 imaging software (PerkinElmer), and deconvolved using Volocity 4 (PerkinElmer). Shown are DIC images (left), 488 nm (TgVTC2) and 633 nm (GRA2, TgVP1 or TgCPL) channels and a composite image (right) including the DNA detection reagent DAPI. All images are identical in scale, and a 5 µm bar is indicated.