Abstract
Waardenburg syndrome (WS) is a genetic disorder characterized primarily by depigmentation of the skin and hair, heterochromia of the irides, sensorineural deafness, and sometimes by dystopia canthorum, and Hirschsprung disease. WS presents a large clinical and genetic heterogeneity. Four different types have been individualized and linked to 5 different genes. We report 2 cases of WS type II and 1 case of WS type IV from Lebanon and Syria. The genetic studies revealed 2 novel mutations in the MITF gene of the WS type II cases and 1 novel homozygous mutation in the EDNRB gene of the WS type IV case. This is the first molecular study of patients from the Arab world. Additional cases will enable a more detailed description of the clinical spectrum of Waardenburg syndrome in this region.
Key Words: Autosomal dominant, Autosomal recessive, Gene, Lebanon, Mutation, Syria, Waardenburg syndrome
Introduction
Waardenburg syndrome (WS) is a genetic disorder characterized by several of the following features: white forelocks or early graying of the hair, patchy depigmentation of the skin, heterochromia of the irides, sensorineural deafness, synophrys, and a high nasal bridge. Some patients might also have dystopia canthorum, Hirschsprung disease and neurological abnormalities.
Four WS types have been individualized on clinical grounds [Read and Newton, 1997]. Type I (WS1; OMIM 193500) is characterized by dystopia canthorum, deafness in 60% of patients, and the distinctive facial features of WS, mainly a high nasal bridge, synophrys, and hypoplasia of the alae nasi [Newton, 1990]. In type II (WS2; OMIM 193510) there is no dystopia canthorum and over 80% of patients have deafness, and 47% heterochromia iridum [Newton, 1990; Liu et al., 1995]. Type III (WS3) or Klein-Waardenburg syndrome (OMIM 148820) is a severe form of type I presenting with skeletal abnormalities. Type IV (WS4) or Shah-Waardenburg syndrome (OMIM 277580) is characterized by the association of WS and Hirschsprung disease. Type II appears to be a specific defect of neural crest-derived melanocytes, whereas type I, III, and IV represent failures of several neural crest derivatives.
Beside its clinical heterogeneity, WS also presents a genetic allelic or non-allelic heterogeneity with autosomal dominant or recessive inheritance, depending on the gene involved. WS1 is caused by loss-of-function mutations in the PAX3 (paired box 3) gene [Baldwin et al., 1992; Tassabehji et al., 1992]. WS2 is a heterogeneous group due in part to mutations in the MITF (microphthalmia-associated transcription factor) [Tassabehji et al., 1994] or SOX10 (SRY (sex-determining region Y)-box 10) [Bondurand et al., 2007] genes. WS3 is caused by mutations in PAX3 [Hoth et al., 1993], some patients being homozygous [Zlotogora et al., 1995]. In WS4, three disease-causing genes have been identified so far: EDNRB encoding the endothelin-B receptor [Puffenberger et al., 1994], EDN3 encoding an endothelin receptor ligand [Edery et al., 1996; Hofstra et al., 1996] and SOX10 [Pingault et al., 1998]. Mutations in the EDNRB and EDN3 genes are inherited in an autosomal recessive manner in most cases, with patients carrying homozygous mutations manifesting WS4, but individuals being heterozygous for mutations in either gene may occasionally present with 1 or more features of the disease in some families. SOX10 mutations are inherited in an autosomal dominant manner, and specific mutations (particularly involving the last coding exon) manifest as a more severe WS4 variant with neurologic findings, characterized by peripheral demyelinating neuropathy and central dysmyelinating leukodystrophy [Inoue et al., 2004].
Types I and II are the most common types of the syndrome, type IV being less frequent, and type III very rare. The highly variable presentation of the syndrome makes it difficult to arrive at precise figures for its prevalence. Overall, WS affects between 1 in 40,000 and 1 in 20,000 people [Read and Newton, 1997; Pingault et al., 2010]. About 1 in 30 students in schools for the deaf have Waardenburg syndrome.
Here, we report 3 cases of WS in Lebanese and Syrian families with novel mutations.
Methods
Clinical Investigations
Three unrelated families, 2 from Lebanon and 1 from Syria, with clinical criteria of Waardenburg syndrome were investigated. The affected individuals were examined extensively, including a thorough clinical evaluation, routine blood tests, and molecular analysis.
Molecular Studies
Molecular studies of the MITF, SOX10, EDN3, and EDNRB genes were undertaken. Informed consent was obtained from each patient. EDTA blood samples were collected for genetic studies, and DNA was extracted from lymphocytes by standard methods.
PCR Amplification. The coding exons of MITF (M isoform), EDN3, EDNRB and SOX10 were analysed by direct sequencing of the PCR products to search for point mutations (SOX10: Bondurand et al. [2007]; MITF: adapted from Tassabehji et al. [1994]; EDN3 and EDNRB: primer sequences available upon request). To detect rearrangements and copy number variants, the coding exons of these genes were analysed by quantitative multiplexed fluorescent (QMF) PCR (SOX10: Bondurand et al. [2007]; MITF, EDN3 and EDNRB: primer sequences and protocols available upon request). Both MITF and SOX10 were analysed in the cases of WS2 (families 1 and 2) and EDN3, EDNRB and SOX10 were studied in the WS4 case (family 3). The variations were named based on the EDNRB (GenBank NM_001122659.1) and the MITF-M isoform (GenBank NM_000248.2) cDNAs, and were tested in 120 control chromosomes.
In silico Analysis. Effects of the sequence variation were predicted by using different software: PolyPhen (http://genetics.bwh.harvard.edu/pph/), Jpred3 (http://www.compbio.dundee.ac.uk/www-jpred/), Alamut v4.0.1 Software (Interactive Biosoftware, France), NetGene2 (http://www.cbs.dtu.dk/services/NetGene2/), and BDGP (http://www.fruitfly.org/seq_tools/splice.html). The strengths of 5′ splice junctions were estimated according to the MaxEnt program (http://genes.mit.edu/burgelab/maxent/Xmax entscan_scoreseq.html) [Yeo and Burge, 2004]. For 5′ splice sites a log-odd ratio (MaxEnt score) is assigned to a 9-mer. The higher the score, the higher the probability that the sequence is a true splice site.
Results
Clinical Findings
Clinical findings of the 3 patients are summarized in table 1.
Table 1.
Clinical and molecular findings in the 3 patients
| Patient 1 | Patient 2 | Patient 3 | |
|---|---|---|---|
| Age at diagnosis (years) | 37 | 28 | 1 |
| Dystopia canthorum | − | − | − |
| White forelock | + | + | + |
| Sensorineural hearing loss | + | + | + |
| Iris pigmentation abnormality | + | + | + |
| Skin hypopigmentation | + | − | + |
| Hirschsprung disease | − | − | + |
| Gene, mutation | MITF c.33+5G>C (intron 1) heterozygous | MITF c.710+5G>T (intron 7) heterozygous | EDNRB p. Ser342Ile homozygous |
Family 1
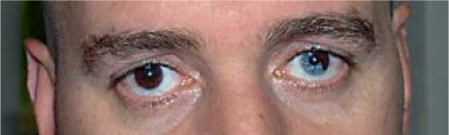
A 37-year-old Syrian male presented for genetic counselling concerning his congenital deafness. Examination revealed skin discoloration with hypopigmented spots on his arms and back of hands. He also had white tufts in his moustache and beard. Inner canthal distance was 30 mm, outer canthal distance was 100 mm, and interpupillary distance was 65 mm, values not suggestive of dystopia canthorum. Segmental heterochromia of the left iris, a high nasal root, and mild synophrys were noted as well (fig. 1).
Fig. 1.
Patient 1. Note the segmental heterochromia.
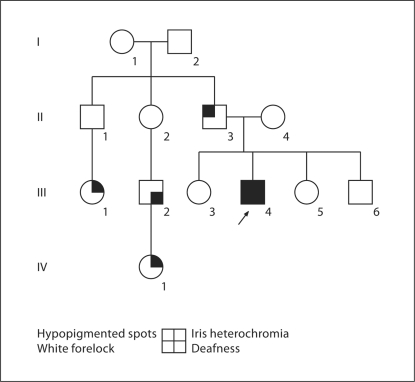
The father of the propositus also had hypopigmented spots and his female first cousin had iris heterochromia. He had another deaf male first cousin, whose daughter had iris heterochromia (fig. 2). There was no family history of Hirschsprung disease or upper limb abnormalities. On the basis of the clinical features and the W index (Waardenburg index) of 1.52, WS2 was suspected and genetic analysis was performed.
Fig. 2.
Pedigree of the family of patient 1 (arrow).
Family 2
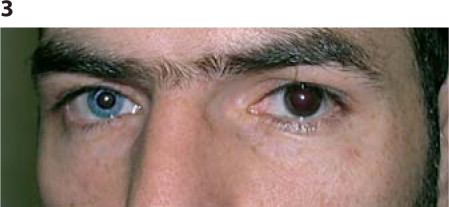
A 28-year-old Lebanese male presented for genetic counselling concerned by his deafness. Examination revealed a high nasal root, bushy eyebrows, synophrys, and a hypochromic blue iris on the right side. There was no dystopia canthorum (fig. 3). His fundus exam was normal. He remembered having a white hair tuft that disappeared after the first few years of life; there was neither hypoplasia of the nasal bone, nor a short retropositioned maxilla. He recalls his mother as the only family member having a white forelock at a young age. WS2 was suspected and genetic analysis was pursued.
Fig. 3.
Patient 2. Note the bushy eyebrows, the synophrys, and the heterochromia.
Family 3
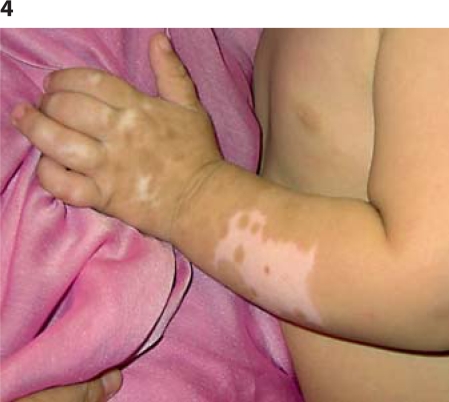
The propositus is the first child of consanguineous Lebanese parents. Hirschsprung disease was diagnosed at 20 days of age and treated with 15-cm resection of the left colon. Bilateral sensorineural deafness was diagnosed by auditory evoked potential at 6 months. When seen by us, he was 15 months old. Clinical examination revealed a white forelock, bilateral hypopigmented blue irises, and hypopigmented skin patches over the forearms (fig. 4). He had no cardiac or abdominal anomalies on ultrasound. WS4 was suspected and a genetic analysis of the baby and both of his parents was performed.
Fig. 4.
Patient 3. Note the depigmented area of the arm.
Molecular Findings
The exploration of the entire coding sequences of SOX10 and MITF (for WS2) or SOX10, EDNRB and EDN3 (for WS4) allowed the identification of likely causative mutations in all 3 families (table 1). The two WS2 patients were found to carry heterozygous MITF intronic substitutions in donor splice sites: c.33+5G>C in intron 1 for patient 1 (fig. 5), and c.710+5G>T in intron 7 for patient 2 (fig. 6). In the young boy of family 3, a single nucleotide substitution was found in homozygous state in exon 5 of the EDNRB gene (c.1025G>T) (fig. 7). This variation predicts the replacement of a serine by an isoleucine at amino acid position 342 of the protein: p. Ser342Ile. His parents were both heterozygous for the substitution, consistent with the consanguinity in this family.
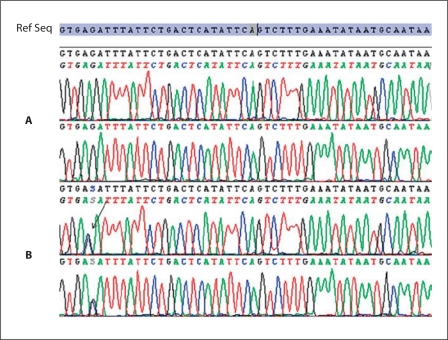
Fig. 5.
Sequencing of exon 1 of the MITF gene. The black arrow shows the position of the mutation. The reference sequence is shown at the top. A Sequences of a healthy individual. B Sequences of the patient heterozygous for the c.33+5G>C mutation.
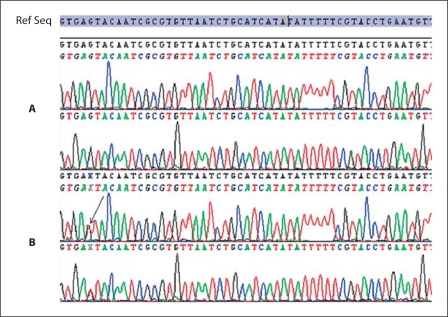
Fig. 6.
Sequencing of exon 7 of the MITF gene. The black arrow shows the position of the mutation. The reference sequence is shown at the top. A Sequences of a healthy individual. B Sequences of the patient heterozygous for the c.710+5G>T mutation.
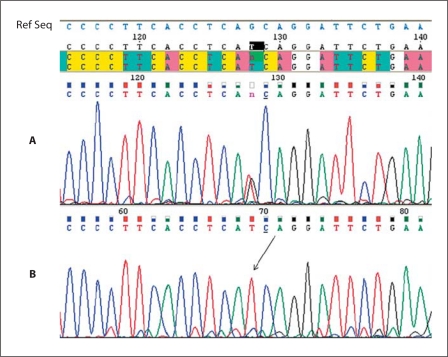
Fig. 7.
Sequencing of exon 5 of the EDNRB gene. The black arrow shows the position of the mutation. The reference sequence is shown in light blue at the top. A Sequence of the patient's father heterozygous for the mutation. B Sequence of the patient homozygous for the mutation.
The 3 sequence variations were absent in 120 control chromosomes.
In silico Results
NetGene2 predicts a complete loss of the donor sites for both mutations c.33+5G>C and c.710+5G>T. BDGP Splice Site Prediction predicts a splicing score of 93% and 91% for the wild-type sequences of c.33+5G>C and c.710+5G>T, respectively, but it predicts a splicing score of 45% for the mutant sequence of c.33+5G>C and a complete loss of the donor site for the mutant sequence of c.710+5G>T. MaxEntScan which evaluates 5′ splice site strengths, calculates for c.33+5G>C a score of 9.22 for the wild-type sequence and a score of 5.49 for the mutant one. It calculates for c.710+5G>T a score of 8.93 for the wild-type sequence and a score of −1.72 for the mutant one; in general, the typical MaxEnt 5′ splice site score is 11.81. PolyPhen, a tool which predicts the possible impact of an amino acid substitution on the structure and function of a human protein, predicts that the p. Ser342Ile mutation in EDNRB gene is possibly damaging.
Discussion
Waardenburg syndrome affects all ethnic groups and both sexes equally. Its precise frequency in the Arab world remains unknown. Only 3 Tunisian patients [Attié et al., 1995; Bonnet et al., 1996] and 1 Omani baby boy [Abdulrazzaq, 1989] have been diagnosed with WS4. No molecular analyses were performed on these patients.
We have been able to study 3 patients with WS from Lebanon and Syria. Overall, their phenotype was in accordance with the literature.
Patients 1 and 2 had WS2. Marked clinical variability between but also within each family was noted, which highlights the importance of modifying factors in WS. Both patients had variations in the MITF gene. About 20 different mutations have been described in this gene [Pingault et al., 2010]. They include splicing alterations and truncating mutations located throughout the gene, as well as a few missense mutations in functional domains of the protein. Our two WS2 patients had a nucleotide substitution in position +5 of the donor consensus site. These substitutions likely cause aberrant splicing, because they occur within the U1 snRNP recognition sequence and replace the conserved guanine at position +5 [Cartegni et al., 2002] with a cytosine and a thymine. Both mutations were absent in 120 control chromosomes, strongly suggesting linkage to the phenotype. In both cases, family members other than the propositus were not available for molecular studies. The position of these 2 substitutions immediately adjacent to the 5′ donor splice site suggested they might disrupt 5′ splice site function and result in the skipping of exons from the mature mRNA leading to truncated proteins. In order to evaluate this hypothesis, the sequences were analyzed with several splice-prediction programs. These showed a strong diminution of splice efficiency for the c.33+5G>C mutation, and a complete loss of the donor site for the c.710+5G>T mutation, which would be expected to be pathogenic. Software predicted the latter mutation may induce use of a cryptic donor site located in exon 7 that would result in a frameshift and premature stop codon.
The WS4 patient had Hirschsprung disease in addition to the symptoms mentioned above. This syndrome is caused by abnormal migration or differentiation of neural crest cells during embryonic development. The enteric nervous system is derived from neural crest cells that reach the foregut and migrate distally along the gut before differentiating. The association of WS features (deafness and depigmentation) with Hirschsprung disease (intestinal aganglionosis), and the involvement of several genes in this syndromic association, indicate common genetic pathways between both lineages. The analysis of the EDNRB gene showed an abnormality in this consanguineous Lebanese family. About 20 different EDNRB mutations have been described to date in WS [Pingault et al., 2010], including truncating and missense mutations. The predicted p. Ser342Ile missense mutation lies in a transmembrane domain. Although involvement of missense variations is always difficult to assess, this serine is conserved among all species and between the 2 members of the protein family, endothelin receptors A and B. Its replacement by an isoleucine is expected to change polarity and steric hindrance. The mutation has a Grantham score of 142 and is predicted as possibly damaging by PolyPhen. In addition, the mutation is absent in 120 control chromosomes. All these observations strongly support that this mutation causes the observed pathogenicity.
In conclusion, we report for the first time the molecular study of 3 families originating from Lebanon and Syria affected by Waardenburg syndrome. We anticipate that other reports of similar cases in Middle East will enable further delineation of the clinical and molecular spectrum of Waardenburg syndrome.
References
- Abdulrazzaq YM. Waardenburg's syndrome with long segment Hirschsprung's disease in an Omani family. Emirates Med J. 1989;7:26–29. [Google Scholar]
- Attié T, Till M, Pelet A, Amiel J, Edery P, et al. Mutation of the endothelin-receptor B gene in Waardenburg-Hirschsprung disease. Hum Mol Genet. 1995;4:2407–2409. doi: 10.1093/hmg/4.12.2407. [DOI] [PubMed] [Google Scholar]
- Baldwin CT, Hoth CF, Amos JA, da-Silva EO, Milunsky A. An exonic mutation in the HuP2 paired domain gene causes Waardenburg's syndrome. Nature. 1992;355:637–638. doi: 10.1038/355637a0. [DOI] [PubMed] [Google Scholar]
- Bondurand N, Dastot-Le Moal F, Stanchina L, Collot N, Baral V, et al. Deletions at the SOX10 gene locus cause Waardenburg syndrome types 2 and 4. Am J Hum Genet. 2007;81:1169–1185. doi: 10.1086/522090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet JP, Till M, Edery P, Attie T, Lyonnet S. Waardenburg-Hirschsprung disease in two sisters: a possible clue to the genetics of this association? Eur J Pediatr Surg. 1996;6:245–248. doi: 10.1055/s-2008-1066521. [DOI] [PubMed] [Google Scholar]
- Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- Edery P, Lyonnet S, Attie T, Amiel J, Pelet A, et al. Mutation of the endothelin-3 gene in the Waardenburg-Hirschsprung disease (Shah-Waardenburg syndrome) Nat Genet. 1996;12:442–443. doi: 10.1038/ng0496-442. [DOI] [PubMed] [Google Scholar]
- Hofstra RMW, Chakravarti A, Meijers C, Buys CHCM, Osinga J, et al. A homozygous mutation in the endothelin-3 gene associated with a combined Waardenburg type 2 and Hirschsprung phenotype (Shah-Waardenburg syndrome) Nat Genet. 1996;12:445–447. doi: 10.1038/ng0496-445. [DOI] [PubMed] [Google Scholar]
- Hoth CF, Milunsky A, Lipsky N, Sheffer R, Clarren SK, et al. Mutations in the paired domain of the human PAX3 gene cause Klein Waardenburg syndrome (WS-III) as well as Waardenburg syndrome type I (WS-I) Am J Hum Genet. 1993;52:455–462. [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Khajavi M, Ohyama T, Hirabayashi S, Wilson J, et al. Molecular mechanism for distinct neurological phenotypes conveyed by allelic truncating mutations. Nat Genet. 2004;36:361–369. doi: 10.1038/ng1322. [DOI] [PubMed] [Google Scholar]
- Liu XZ, Newton VE, Read AP. Waardenburg syndrome type 2: phenotypic features and diagnostic aspects. Am J Med Genet. 1995;55:95–100. doi: 10.1002/ajmg.1320550123. [DOI] [PubMed] [Google Scholar]
- Newton V. Hearing loss and Waardenburg's syndrome: implications for genetic counselling. J Laryngol Otol. 1990;104:97–103. doi: 10.1017/s002221510011196x. [DOI] [PubMed] [Google Scholar]
- Pingault V, Bondurand N, Kuhlbrodt K, Goerich DE, Préhu MO, et al. SOX10 mutations in patients with Waardenburg-Hirschsprung disease. Nat Genet. 1998;18:171–173. doi: 10.1038/ng0298-171. [DOI] [PubMed] [Google Scholar]
- Pingault V, Ente D, Dastot-Le Moal F, Goossens M, Marlin S, et al. Review and update of mutations causing Waardenburg syndrome. Hum Mutat. 2010;31:391–406. doi: 10.1002/humu.21211. [DOI] [PubMed] [Google Scholar]
- Puffenberger EG, Hosoda K, Washington SS, Nakao K, deWit D, et al. A missense mutation of the endothelin-B receptor gene in multigenic Hirschsprung's disease. Cell. 1994;79:1257–1266. doi: 10.1016/0092-8674(94)90016-7. [DOI] [PubMed] [Google Scholar]
- Read AP, Newton VE. Waardenburg syndrome. J Med Genet. 1997;34:656–665. doi: 10.1136/jmg.34.8.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassabehji M, Read AP, Newton VE, Harris R, Balling R, et al. Waardenburg's syndrome patients have mutations in the human homologue of the Pax-3 paired box gene. Nature. 1992;355:635–636. doi: 10.1038/355635a0. [DOI] [PubMed] [Google Scholar]
- Tassabehji M, Newton VE, Read AP. Waardenburg syndrome type 2 caused by mutations in the human microphthalmia (MITF) gene. Nat Genet. 1994;8:251–255. doi: 10.1038/ng1194-251. [DOI] [PubMed] [Google Scholar]
- Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- Zlotogora J, Lerer I, Bar-David S, Ergaz Z, Abeliovich D. Homozygosity for Waardenburg syndrome. Am J Hum Genet. 1995;56:1173–1178. [PMC free article] [PubMed] [Google Scholar]