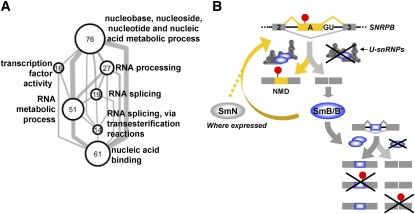
Figure 7.
Model for the role of core snRNP proteins in AS regulation. (A) Enriched GO terms (P < 0.005 and FDR < 0.1) annotating genes containing exons affected by SmB/B′ knockdown (≥30% more skipping) are represented as a network of gene sets. Each node represents the set of genes annotated with the indicated GO term. Node size is proportional to the number of genes annotated by the term (indicated by the node label), and edge thickness is proportional to the number of genes in common between the sets. (B) Model summarizing our data. Levels of SmB/B′ are regulated through homeostatic feedback acting via AS-NMD. The related tissue-restricted Sm protein SmN also cross-regulates SmB/B′ (see the Discussion). SmB/B′ levels, through effects on functional snRNP concentrations, also promote inclusion of alternative exons in many other genes, a subset of which also regulate transcript levels via NMD.