Abstract
Principal component analysis (PCA) and factor analysis (FA) are often used to uncover genetic factors that contribute to complex disease phenotypes. The purpose of such an analysis is to distill a genetic signal from a large number of correlated phenotype measurements. That signal can then be used in genetic analyses (e.g. linkage analysis), presumably leading to greater success at finding genes than one would achieve with any one raw trait. Although both PCA and FA have been used this way, there has been no comparison of their performance in the literature. We compared the ability of these two procedures to extract unobserved underlying genetic components from complex simulated data on nuclear families. We first simulated 7 underlying genetic and environmentally determined traits. Then we derived two sets of 50 complex (observed) traits using algebraic combinations of the underlying components. We next performed PCA and FA on the complex traits. We assessed two aspects of the performance of the methods: 1) ability to detect the underlying genetic components; 2) whether the methods worked better when applied to raw traits or to residuals (after regressing out significant environmental covariates). Our results indicate that both methods behave similarly in most cases, although FA generally produced factors that had stronger correlations with the underlying traits. We also found that using residuals in PCA or FA analyses greatly increased the probability that the PCs or factors detected common genetic components instead of common environmental factors, except if there was statistical interaction between genetic and environmental factors.
Keywords: multivariate analysis, PCA, factor analysis, quantitative traits
INTRODUCTION
Numerous studies over the past several decades indicate that genes contribute to the development of complex diseases such as osteoporosis, obesity, and diabetes. Many risk factors for these diseases (such as bone mineral density, body fat, glucose levels) have been shown to be moderately to highly heritable. In recent years, many studies have suggested that a majority of these highly heritable traits (risk factors) are governed by a set of common genes (i.e. pleiotropy, defined as when two or more phenotypes are co-regulated by a common gene or a common sets of genes) [Deng, et al. 2006; Hegele 1997; Li, et al. 2002; Mitchell, et al. 1996]. One piece of evidence in support of the above hypothesis is that bivariate linkage analyses of some of these traits revealed stronger linkage signals than were obtained from univariate linkage analysis of each trait separately [Devoto, et al. 2005; Li, et al. 2006; Livshits, et al. 2004; Martin, et al. 2004].
Conventional measurements of these complex disease-related phenotypes produce many intercorrelated phenotypes. For example, Aldridge et al. measured multiple regions of the brain using 3-D MRI, resulting in more than 30 different landmarks and corresponding phenotype measurements [Aldridge, et al. 2005]. Wang et al. in 2007 generated more than 100 different correlated bone traits including bone mineral density (BMD) and bone geometry measures produced by Dual-Energy X-Ray Absorptiometry (DXA) and peripheral Quantitative Computed Tomography (pQCT) [Wang, et al. 2007a] and [Wang, et al. 2007b]. In studies such as these, it is possible that there might be a relatively small number of factors (both genetic and environmental) involved in certain metabolic pathways that contribute to variation in an underlying cluster of phenotypes. Identification of these common factors and elucidation of their molecular basis should contribute to a better understanding of and possible treatment for some complex diseases.
It is well-known that bivariate and tri-variate genetic analyses are computationally intensive. Genetic analyses of more than three traits are beyond our current computational ability. Therefore, multivariate analysis might be an alternative yet effective solution to identify common genetic factors that affect multiple traits. Principal component analysis (PCA) or factor analysis (FA) can be used to extract these underlying factors, which can then be used as phenotypes in a genetic linkage or association study. Both PCA and FA involve a mathematical procedure that transforms a number of (possibly) correlated variables into a (smaller) number of uncorrelated (PCA) or correlated (FA) variables called principal components or factors. During the PCA/FA extraction, the shared variance of a variable is partitioned from its unique variance and error variance to reveal the underlying factor/PC structure. Only the shared variance appears in the solution. So it is reasonable to hypothesize that these two methods have the potential to classify phenotypic variation into independent/dependent components that may amplify or purify genetic signals and hence be used to dissect genetic networks regulating complex biological systems. Although the two analyses can be very similar, there exist some subtle differences between them. First, in PCA we assume that all variability in an item should be used in the analysis, while in factor analysis, we define a priori the number of factors that we want to extract, and the extracted axes will be scaled to the variance along the new improved axes. Second, in PCA the goal is to account for as much of the total variance in the variables as possible, while FA is trying to explain the covariances or correlations among the variables. Finally, people tend to use PCA to reduce the data into a smaller number of components, while they use FA to understand what constructs underlie the data.
Since 2001, ten groups of investigators that we are aware of have published articles that used multivariate analysis methods in an attempt to dissect the genetic and environmental basis for complex diseases, such as osteoporosis, metabolic syndrome, and asthma. Seven of these groups applied PCA [Chase, et al. 2002; Guo, et al. 2005; Hakulinen, et al. 2006; Karasik, et al. 2004; Lin, et al. 2005; Musani, et al. 2006; Peacock, et al. 2004], while the other three used FA [Austin, et al. 2004; Holberg, et al. 2001; Lee, et al. 2004]. In addition, 7 groups used raw phenotypes directly as the input variables, one group used raw traits but performed analysis by gender and generation [Karasik, et al. 2004], and the last two groups used residuals (after adjustment for significant covariates) [Austin, et al. 2004; Lin, et al. 2005]. The goals of the 10 groups also differed: one group used multivariate analysis for phenotype clustering/classification, by which it developed composite index scores summarizing characteristics of raw traits from different skeletal sites [Lee, et al. 2004]. The remaining nine groups all focused on exploring the underlying genetic/environmental basis of composite traits (that is, principal components or factors) derived from PCA or FA. Among these nine groups, two reported genetic or environmental correlations between composite traits and some well-defined real (observed) phenotypes [Guo, et al. 2005; Hakulinen, et al. 2006]; two reports focused exclusively on heritability estimation for composite traits [Austin, et al. 2004; Lin, et al. 2005]; and three reports concentrated on the association (or linkage) between these composite traits and QTLs (Quantitative Trait Loci) [Holberg, et al. 2001; Musani, et al. 2006; Peacock, et al. 2004]. The final two papers performed both heritability estimation and association/linkage analysis for composite phenotypes [Chase, et al. 2002; Karasik, et al. 2004].
However, many statistical issues remain unaddressed by these reports. First, the selection of either PCA or FA seems arbitrary; none of the groups justified why they chose one instead of the other. Consequently we decided to evaluate the performance of these two approaches. In particular, we wanted to assess which method is better able to detect underlying genetic factors, with the idea that such factors should be more powerful traits to use in genetic linkage and association analyses than the original traits. Second, most reports used raw traits as input variables, but a few used residuals after regressing out some important environmental factors. Does analysis of residuals significantly improve the ability of PCA or FA methods to detect underlying genetic components? No direct comparisons to answer this question have been reported.
The goal of our study is to address the two issues named in the preceding paragraph. We do this by simulating underlying environment and genetic traits, then observable complex traits that are functions of the underlying traits. We apply both PCA and FA to the complex traits, and measure the ability to recover the underlying genetic components. In all cases, the presumed purpose of using FA or PCA is to distill a genetic signal. That signal can then be used in genetic analyses (e.g. linkage analysis), presumably leading to greater success at finding genes than one would achieve with any one raw trait. Thus, the outcome of the FA or PCA that we want to measure in this study is the correlation between the factors or components and the underlying genetic signals (which we know because we simulated the data – see methods). Specifically, our outcome measure is the percentage of variance (R2) of each of the first three factors/components that is explained by each of the 7 underlying phenotypes. Methods that result in higher correlations with genetic components (as measured by percentage of variance explained) are judged to be better than methods that produce components with lower correlations. Note that this outcome measure is not necessarily related to any traditional FA or PCA outcome measure such as ability to recover the correct model (since the correct model may involve non-linear functions), ability to maximize the percent of overall variance explained, etc. Our choice of outcome measure is directly related to the ultimate goals of using PCA or FA in this context, which are to “discover” a hidden genetic component that can then be used as a phenotype in a gene-mapping study. Such a gene mapping study will have the greatest chance of success if the component or factor that we discover has a high correlation with a single genetic effect. Thus our sole outcome measure is correlations between the components and the underlying genes.
METHODS
STUDY DESIGN
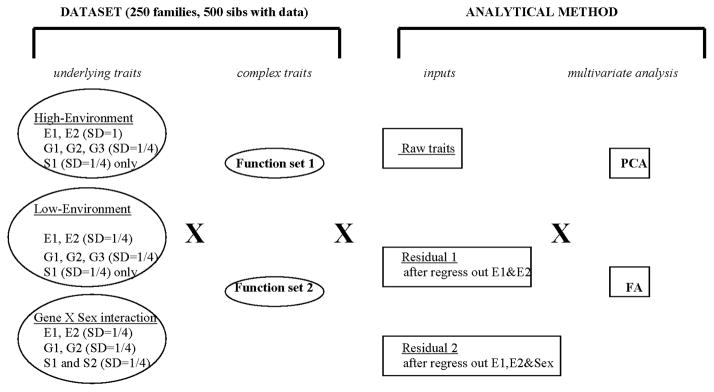
Our overall study design is illustrated in Figure 1. Three datasets of underlying traits were generated by simulation. The underlying traits are E1 and E2 (environmental effects, which can be treated as either observed or unobserved), G1, G2, and G3 (unobserved genetic components), and S1 and S2 (unobserved genetic components with sex-dependent effects). Details on how the traits were simulated are given later in this section. The differences among the three datasets are in the variances of the environmental traits (E1 and E2) and the inclusion or exclusion of S2 (a gene by sex interaction trait). For each of these three datasets of underlying traits, two sets of complex phenotypes were created using arbitrary algebraic functions of the underlying traits. There are 50 complex traits in each of the two function sets. Set 1 involves somewhat simpler algebraic combinations of traits than set 2 (details in table II and III). The seven underlying traits represent the environmental or/and genetic determinants that influence population variation of observable traits, which are in turn represented by the sets of 50 complex traits. Using these complex traits, we created three different inputs for further multivariate analysis: raw traits, residuals model 1 (after regressing out E1 and E2); and residuals model 2 (after regressing out E1, E2 and sex). Finally, we performed both PCA and FA on each of the dataset × function set × residual combinations (18 scenarios in total). (Figure 1). We performed each of these analyses on a total of six replicates of the underlying trait datasets. All datasets and replicates used the same sample size, pedigree structure and gender ratio (male/female =50/50). Each aspect of the study design is described in more detail below.
Fig 1.
Blueprint for study design
Table II.
Underlying phenotypes picked up by factors and principal components for function set 1
| A: Low Environment Model | ||||||
|---|---|---|---|---|---|---|
| Raw Traits | Residual 1 | Residual 2 | ||||
| E1 and E2 regressed out | E1, E2 &Sex regressed out | |||||
| Factor (R2) | PC (R2) | Factor (R2) | PC (R2) | Factor (R2) | PC (R2) | |
| First | S1 (0.92)* | E1+S1(0.23,0.49) | S1 (0.95) | S1 (0.79) | S1 (0.65) | G2+S1(0.20,0.48) |
| Second | E1 (0.92) | G2+G3(0.36,0.26) | G2 (0.94) | G2+G3(0.30,0.45) | G2 (0.93) | G2+G3(0.24,0.45) |
| Third | E2 (0.87) | E1(0.65) | G3(0.91) | G1 (0.54) | G3(0.92) | G1 (0.53) |
| B: High Environment Model | ||||||
|---|---|---|---|---|---|---|
| Raw Traits | Residual 1 | Residual 2 | ||||
| E1 and E2 regressed out | E1, E2&Sex regressed out | |||||
| Factor (R2) | PC (R2) | Factor (R2) | PC (R2) | Factor (R2) | PC (R2) | |
| First Component | E1 (0.94) | E1+E2(0.34,0.26) | S1 (0.92) | S1 (0.68) | S1 (0.59) | G2+S1(0.31,0.30) |
| Second Component | E2 (0.90) | E1+E2(0.40,0.52) | G2 (0.93) | G2+G3(0.37,0.27) | G2 (0.94) | G3+S1(0.24,0.45) |
| Third Component | S1 (0.91) | E1+G2(0.20,0.38) | G3(0.87) | G1+G3(0.59,0.28) | G3(0.88) | G1+G3(0.61,0.21) |
| C: Gene by Environment Model | ||||||
|---|---|---|---|---|---|---|
| Raw Traits | Residual 1 | Residual 2 | ||||
| E1 and E2 regressed out | E1, E2&Sex regressed out | |||||
| Factor (R2) | PC (R2) | Factor (R2) | PC (R2) | Factor (R2) | PC (R2) | |
| First Component | S2 (0.79) | S1+S2(0.52,0.72) | S2 (0.91) | S1+S2(0.52,0.83) | S2 (0.23) | G2+S1(0.26,0.18) |
| Second Component | S1 (0.87) | G1+G2(0.18,0.30) | G3 (0.85) | G2+S1(0.11,0.23) | S1 (0.60) | S1(0.34) |
| Third Component | E1 (0.66) | E1(0.77) | G2 (0.92) | G1+G2(0.28,0.34) | G2 (0.93) | G1+G2(0.41,0.22) |
R2 indicates percent of variation in the underlying trait that is explained by the factor or principal component. Estimates of R2 were based on data from 250 independent siblings (one from each family).
Table III.
Underlying phenotypes picked up by factors and principal components for function set 2
| A: Low Environment Model | ||||||
|---|---|---|---|---|---|---|
| Raw Traits | Residual 1 | Residual 2 | ||||
| E1 and E2 regressed out | E1, E2 &Sex regressed out | |||||
| Factor (R2) | PC (R2) | Factor (R2) | PC (R2) | Factor (R2) | PC (R2) | |
| First Component | S1 (0.92)* | E1+G1(0.43,0.30) | G1 (0.92) | G1 (0.76) | G1 (0.93) | G1 (0.84) |
| Second Component | E1 (0.89) | S1(0.70) | S1 (0.94) | S1 (0.75) | S1 (0.57) | G2+S1(0.44,0.20) |
| Third Component | G1 (0.91) | G2+G3(0.26,0.46) | G3(0.91) | G2+G3(0.35,0.48) | G3 (0.91) | G3+S1(0.42,0.27) |
| B: High Environment Model | ||||||
|---|---|---|---|---|---|---|
| Raw Traits | Residual 1 | Residual 2 | ||||
| E1 and E2 regressed out | E1, E2&Sex regressed out | |||||
| Factor (R2) | PC (R2) | Factor (R2) | PC (R2) | Factor (R2) | PC (R2) | |
| First Component | E1 (0.93) | E1 (0.81) | G1 (0.89) | G1 (0.80) | G1 (0.90) | G1(0.82) |
| Second Component | E2 (0.93) | E2(0.71) | S1 (0.93) | G2+S1(0.32,0.51) | S1 (0.68) | G2+S1(0.44,0.25) |
| Third Component | S1 (0.92) | G2+S1(0.24,0.22) | G3 (0.91) | G3+S1(0.55,0.25) | G3 (0.90) | G3+S1(0.51,0.24) |
| C: Gene by Environment Model | ||||||
|---|---|---|---|---|---|---|
| Raw Traits | Residual 1 | Residual 2 | ||||
| E1 and E2 regressed out | E1, E2&Sex regressed out | |||||
| Factor (R2) | PC (R2) | Factor (R2) | PC (R2) | Factor (R2) | PC (R2) | |
| First Component | S2 (0.81) | S1+S2(0.61,0.80) | G1 (0.92) | S1+S2(0.56,0.81) | G1 (0.92) | G1(0.71) |
| Second Component | S1 (0.93) | E1+G1(0.36,0.50) | S2 (0.80) | G1(0.86) | S2 (0.31) | G1+G2(0.17, 0.22) |
| Third Component | E1 (0.88) | E1+S1(0.32,0.11) | S1 (0.93) | G2+S1(0.23,0.20) | S1 (0.60) | S1+S2(0.19,0.15) |
R2 indicates percent of variation in the underlying trait that is explained by the factor or principal component. Estimates of R2 were based on data from 250 independent siblings (one from each family).
SIMULATIONS
We first simulated 250 nuclear families with two parents and two offspring within each family. Parental alleles were chosen assuming Hardy-Weinberg equilibrium, and alleles were transmitted to children using standard Mendelian rules. We then simulated phenotypes E1, E2, G1, G2, G3, S1, and S2, (Table I) for the offspring only, for a total of 500 phenotyped individuals. All of these underlying traits except for E1 and E2 were assumed to be normally distributed conditional on genotype. Because some environmental factors are likely to be similar between siblings, we also allowed for the effect of a shared common environment for E1 and E2 by simulating these two traits based on a bivariate normal distribution with means all equal 1, with standard deviation equal to 1 or 1/4 for different datasets and covariance between the two sibs equal to 0.2 for E1 and 0.1 for E2. Both E1 and E2 can be thought of as either observed or unobserved. If they are unobserved, they cannot be regressed out before applying PCA or FA. But if they are observed, one has a choice of whether to perform PCA or FA on raw traits or on residuals. Traits G1, G2 and G3 are standard simple genetic models in which means differ among genotypes. As can be seen in Table I, the genotypic means and error variances for, G1, G2 and G3 are identical (mean 1.5, 2.5 and 3.5 for genotype aa, Aa and AA respectively and all standard deviations = 1/4); only the allele frequencies of these traits differ. The trait S1 has different genotypic effects in males and females, but the same allele frequency in both sexes, and no interaction between sex and genotype. The trait S2 incorporates sex by genotype interaction.
Table I.
Simulation parameters for 7 underlying phenotypes
| Parameter | Sex-Specific Genotype | E1 | E2 | G1 | G2 | G3 | S1 | S2 |
|---|---|---|---|---|---|---|---|---|
| Mean | ♂-aa | 1 | 1 | 1.5 | 1.5 | 1.5 | 2 | 1.5 |
| ♂-Aa | 1 | 1 | 2.5 | 2.5 | 2.5 | 3 | 2.5 | |
| ♂-AA | 1 | 1 | 3.5 | 3.5 | 3.5 | 4 | 3.5 | |
| ♀-aa | 1 | 1 | 1.5 | 1.5 | 1.5 | 1 | 1 | |
| ♀-Aa | 1 | 1 | 2.5 | 2.5 | 2.5 | 2 | 1 | |
| ♀-AA | 1 | 1 | 3.5 | 3.5 | 3.5 | 3 | 1 | |
| SD | 1 or 1/4 | 1 or 1/4 | 1/4 | 1/4 | 1/4 | 1/4 | 1/4 | |
| P(a) | 0.8 | 0.9 | 0.95 | 0.8 | 0.7 | |||
COMPLEX TRAITS
Based on the above underlying traits, we created two sets of 50 complex traits. The first set of 50 complex traits is algebraic combinations of a subset of the 7 underlying traits plus an error term, which is normally distributed with mean 1 and standard deviation 1 (Supplement table I-A). (The mean of 1 is equivalent to adding a constant to all trait values). The algebraic functions include additive functions of the underlying components as well as multiplicative, reciprocal, and even exponential. We stress that these functions are extremely arbitrary and nonlinear, and that the resulting complex traits have a wide variety of distributions. Our objective in choosing these functions was to reflect the current genetic/epidemiological assumptions about complex traits regarding the effects of underlying genetic/environment factors. For example, we used additive and multiplicative effects as well as their combinations within and/or between underlying genetic and environmental traits. In addition, we included several very complicated functions so that we could assess the ability of PCA and FA to recover underlying traits even from extremely complex traits.
In order to assess even more complex models, we then created another set of 50 complex traits, in which we removed some of the algebraically simpler combinations and substituted more complex ones. These new 50 functions were similar in format to the more complicated ones in the first set of functions (Supplement table I-B). When devising our 50 complex traits for each set, we required that each underlying trait have a similar representation across all 50 complex traits. Each underlying trait appears in approximately 57% of the complex trait formulas, with percentages for the different underlying traits vary from 54% to 60%.
These complex traits represent phenotypes that we could observe or directly measure in reality, such as bone mineral density (BMD), body mass index (BMI), glucose level, and blood pressure. The seven original traits represent underlying genetic or environmental components; they contribute to the true variation of the measured (complex) traits but would not be actually observed or measured (except perhaps E1 and E2).
DATASETS
For each set of functions, we created three different datasets of underlying traits by simulation to evaluate the performance of the multivariate analysis methods. Datasets 1 and 2 use only 6 out of 7 underlying traits: E1, E2, G1, G2, G3, and S1 (see Supplement table I-A, I-B and Figure 1). The only differences between these two datasets are the standard deviations of E1 and E2 as is described above, i.e. the amount of environmental “noise” on top of the genetic signal. For the third dataset, we substituted underlying traits S1 for G3 and S2 for S1. However, we kept the functions the same and set the standard deviation equal 1/4 for E1 and E2. Taking trait C49 in the second set of functions as an example, we used
for datasets 1 and 2, and
for dataset 3.
We designed these three datasets to perform the following comparisons. 1) By comparing analysis results from datasets 1 and 2, we could assess the behavior of the two multivariate analysis methods when the proportion of trait variation that is due to environmental effects changes. 2) By comparing analysis results from datasets 2 and 3, we could evaluate the behavior of the analysis methods with and without the presence of sex by genotype interaction. For simplicity, we will refer to datasets 1, 2 and 3 in the subsequent text as the high-environment dataset, the low-environment dataset, and the gene by sex interaction dataset, respectively. Finally, each of our datasets was replicated six times, so that consistency of results could be assessed.
STATISTICAL ANALYSIS
The input variables for the multivariate analyses were either the 50 complex traits in their original form (raw traits), or residuals of these traits after removing the linear effect of covariates. Two types of residuals were analyzed: 1) residuals after adjusting for E1 and E2; and 2) residuals after adjusting for E1, E2 and sex. Both sets of residuals were derived from each of the 50 continuous traits by multiple regressions after the incorporation of corresponding covariates. To mimic analysis methods that are typically used in real studies, we only considered the linear form of covariates in the multiple regression, although quadratic and other non-linear effects of E1 and E2 are included in our arbitrary functions.
Principal component analysis and factor analysis were both performed in R, [R-Development-Core-Team 2007] using the following options (varimax rotation, covariance matrix = Pearson correlation matrix) together with all other default options. (Commands: princomp and factanal). In FA, we also used the embedded significance-test statistic to check the minimal number of factors needed in the model. Then we use this minimal number in FA as the desired number of output components. For simplicity, we performed comparisons using only the first three PCs or factors; these explained the major proportion of variation (30–50%) across the 50 complex traits.
EVALUATION
We evaluated the ability of each method to detect underlying genetic components by performing univariate regression analyses and regressing each underlying trait on each PC and factor for 250 independent sibs (one from each family). We report the R2 for each of these models, i.e. the percentage of variation in each factor/PC that is explained by each underlying trait. Also for each factor/PC, we report which underlying trait(s) it picks up, as defined by the following rules. If one of the underlying traits’ R2 is at least three-fold higher than the others, we considered this to be the only signal picked up. If the ratio is less than three-fold, we arbitrarily report the two underlying traits with the highest R2 values as the identified signals.
RESULTS
Tables II and III report the underlying traits picked up by the first three factors and principal components for each of our 18 scenarios. They also show the R2s for each factor/PC and underlying trait. We report here the results from the first replicate, with results from all replicates reported in the supplement. The results are highly consistent across replicates in terms of which traits are picked up, although the proportion of variance explained is variable in some cases, especially for PCA. Consistency across replicates is discussed further below.
Tables II and III show that in essentially all of our 18 scenarios principal components and factor analysis were both able to pick out signals of underlying traits from our large set of complex traits, but that factor analysis almost always detected “purer” signals in the sense of finding factors that had a higher R2 with a single underlying trait. For example, for function set 1 (Table II), low environment model, “residual 2,” the three factors correspond to underlying components S1, G2, and G3 with R2s of 0.65, 0.93, and 0.92. For the same scenario, two of the three principal components pick up combinations of underlying traits, and the highest R2 between any of the three principal components and any of the underlying traits is 0.65 for the third principal component and G1.
As mentioned above, the results are very consistent across replicates, particularly for FA. For example, for function set 2, low environment model, “residual 1” (Supplemental table III-B), the first two factors always correspond to G1 and S1, with R2s ranging from 0.84 to 0.93 for G1 and 0.85 to 0.95 for S1. The third factor corresponds to G3 in five out of six replicates, with R2s ranging from 0.90 to 0.94. In principal component analysis of the same scenario, the first two components correspond to G1 and S1 or a combination of the two in all replicates but one, with R2s for G1 ranging from 0.24 to 0.91 and R2s for S1 ranging from 0.30 to 0.83. The third principal component corresponds to a combination of G2 and G3 in five out of six replicates.
We also compared the performance of the three different regression strategies. We found that when using raw traits both PCA and FA are more likely to pick up the environmental factors. This is especially true in the high environment model, as one might expect. For example, in both function sets 1 and 2 (Tables II-B and III-B), for the high environment model with raw traits, the signals captured by the first two factors and the first two principal components are E1 and E2. However, when E1 and E2 were regressed out (“residual 1”), both methods picked up the underlying genetic signals instead. These results suggest that performing a linear regression of environmental factors might be effective in removing even the non-linear effects of environmental factors, but this may depend on the specific set of non-linear functions we used and thus further evaluation is needed.
Finally, our results also indicate that regressing out a covariate (sex, in our example) that has an interaction effect with an underlying trait, substantially decreases the ability of PCA or FA to detect the underlying trait. For example, consider table II part C, in the gene by environment model. For factor analysis, S2 was captured as the first component, with an R2 of 0.79 in the raw trait analysis. But for “residuals 2” (adjusting for E1, E2 and sex), it was captured with an R2 of only 0.23. Similar results were observed for the principal components, and also for both factors and principal components in function set 2 (Table III).
DISCUSSION
Overall, these results show that both PCA and FA are able to perform well at their intended task of “detecting” underlying shared components of the complex traits. According to our results, there is an obvious performance edge to FA, based on its capability of capturing single underlying components with higher R2 in almost all cases, but this could be dependent on the particular set of functions we chose to generate our complex traits and definitely needs further evaluation. Another reason we prefer FA, although not discussed in the results, is that PCA assumes an orthogonal relationship between its PCs, while FA does not. The assumption of independent extracted components may conflict with the true genetic model. For example, bone scientists hypothesize that genes influencing bone size may differ from genes influencing for bone mineral density (BMD) [Klein, et al. 2002; Masinde, et al. 2003]. However these two sets of genes might interact with each other. If we put several bone size and BMD traits together into PCA, it is almost impossible to generate two independent PCs which represent both the set of the bone size genes and BMD genes respectively.
Our results also show that the methods perform better if environmental effects are regressed out, at least if the environmental effects are not interacting with the genetic effects. We only regressed out the linear effects of E1 and E2, even though our complex traits are in fact non- linear functions of E1 and E2. But our results suggest that this linear regression may be effective in removing some of the non-linear effects of environmental correlates as well. We also showed, however, that if the environmental effect is interacting with a gene, regressing out the environment can make it difficult to detect the interacting gene.
Our results regarding environmental covariates have two different practical implications for real studies. First, any covariate that is not observed obviously cannot be regressed out, so one must be aware when applying FA or PCA that the factors/components that are identified could represent underlying environmental factors rather than underlying genetic factors. Second, for any covariate that is observed (e.g. age or sex), we have two choices – we can either regress it out or not. If we regress it out, we improve our results as long as it does not interact with the genetic effects, but we might lose our ability to detect the genetic effects if there is an interaction. Alternatively, if we do not regress it out, we can perform the factor analysis or principal components analysis and then check the correlations of the factors/components with the observed covariate. If we find that we have “discovered” a factor that is highly correlated with our observed covariate, we can just omit that factor from our downstream genetic analyses since we know that it does not correspond to a genetic signal. This may preserve our ability to detect interacting genes, if they exist. We did not, however, explicitly test this approach in this paper.
Our general qualitative results are very robust across all six replicates, and our detailed numerical results are quite consistent as well. For each of our 18 scenarios, FA and PCA tended to pick up exactly the same underlying trait in each replicate. More importantly, the R2s corresponding to the signal showed relatively little variation, especially for FA. Also, when we examined the minor discrepancies, we observed that most of the time, the differences were due to one of the following situations: 1) same underlying traits but different order (for example, we identified E1, S1 and G2 for the first three factors respectively in one replicate and S1, E1 and G2 in the second replicate); 2) we arbitrarily reported in our table only the two strongest signals when two or more similar R2 values were found (refer to methods, selection criteria). In this situation, even minor differences in R2 sometimes resulted in changes in which two signals were strongest. However, the critical point is that none of this variation affects any of our basic results. All our conclusions hold true in each of the 6 replicates.
One criterion that is often used to judge the success of factor analysis or principal components analysis for detecting underlying genetic traits is if the heritabilities of the factors/components are higher than those of the observed traits. Based on our study, we feel that this is not a reliable criterion for success. We found that there was no predictable relationship between the heritability of the factors/components and the heritability of the 50 complex (observed) traits, even when the factors/components were clearly detecting the underlying (unobserved) genetic signals (data not shown). The heritabilities of the factors/components traits were not necessarily higher than those of the observed traits. We believe that the reason for our observation is that the heritabilities of the observed complex traits reflect combined effects of multiple genes, while the heritabilities of our factors/components reflect effects of only one gene at a time. In any downstream genetic analyses (e.g. linkage analysis), what really matters is the heritability attributable to any single locus, so the highest heritability does not necessarily correspond to the highest chance of success.
Certain limitations of this study need to be acknowledged. One of the most important is a more definitive comparison of PCA vs. FA would require simulating marker data as well as phenotypes, and then following up the PCA or FA with a linkage or association analysis to see which factors/components yield the highest power to detect genes. We also did not assess the robustness of the multivariate methods to the violation of independence when family data are used. We calculated our factors and components based on siblings, as is widely done in applications of these methods to genetic data. But our families only contained two siblings, and in larger families the violation of the independence assumption could conceivably cause problems. In addition, the effect of sample size on our results should be studied. We simulated 250 families, or 500 sibs with phenotypic data, which is comparable to real datasets to which these methods have been applied. The stability of our results across replicates shows that this sample size is adequate for detecting underlying genetic effects given our function sets, but it will be important to know whether FA and PCA also perform well for smaller sample sizes and/or smaller genetic effects. Finally, the question of how many factors or components one looks at in this type of study is somewhat controversial. We looked at only three, and some investigators might consider far more. Since we did not look at results for 4th or 5th (or 10th) factors or components, it is probably not appropriate to extrapolate our results to those.
Supplementary Material
Acknowledgments
Dr. Wang was supported by Grants R03-AR050107 and R01-AR049747 (PI Zmuda) from the National Institute of Arthritis and Musculoskeletal Diseases during his PhD training and currently is supported by NIH/NIDCR grant # R01-DE014899 and NIH grant # U01-DE018903 (PI Marazita).
References
- Aldridge K, Kane AA, Marsh JL, Panchal J, Boyadjiev SA, Yan P, Govier D, Ahmad W, Richtsmeier JT. Brain morphology in nonsyndromic unicoronal craniosynostosis. Anatomical Record. Part A, Discoveries in Molecular, Cellular, & Evolutionary Biology. 2005;285(2):690–8. doi: 10.1002/ar.a.20201. [DOI] [PubMed] [Google Scholar]
- Austin MA, Edwards KL, McNeely MJ, Chandler WL, Leonetti DL, Talmud PJ, Humphries SE, Fujimoto WY. Heritability of multivariate factors of the metabolic syndrome in nondiabetic Japanese americans. Diabetes. 2004;53(4):1166–9. doi: 10.2337/diabetes.53.4.1166. [DOI] [PubMed] [Google Scholar]
- Chase K, Carrier DR, Adler FR, Jarvik T, Ostrander EA, Lorentzen TD, Lark KG. Genetic basis for systems of skeletal quantitative traits: principal component analysis of the canid skeleton. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(15):9930–5. doi: 10.1073/pnas.152333099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng FY, Lei SF, Li MX, Jiang C, Dvornyk V, Deng HW. Genetic determination and correlation of body mass index and bone mineral density at the spine and hip in Chinese Han ethnicity. Osteoporosis International. 2006;17(1):119–24. doi: 10.1007/s00198-005-1930-4. [DOI] [PubMed] [Google Scholar]
- Devoto M, Spotila LD, Stabley DL, Wharton GN, Rydbeck H, Korkko J, Kosich R, Prockop D, Tenenhouse A, Sol-Church K. Univariate and bivariate variance component linkage analysis of a whole-genome scan for loci contributing to bone mineral density. European Journal of Human Genetics. 2005;13(6):781–8. doi: 10.1038/sj.ejhg.5201411. [DOI] [PubMed] [Google Scholar]
- Guo Y, Zhao LJ, Shen H, Guo Y, Deng HW. Genetic and environmental correlations between age at menarche and bone mineral density at different skeletal sites. Calcified Tissue International. 2005;77(6):356–60. doi: 10.1007/s00223-005-0181-x. [DOI] [PubMed] [Google Scholar]
- Hakulinen MA, Day JS, Toyras J, Weinans H, Jurvelin JS. Ultrasonic characterization of human trabecular bone microstructure. Physics in Medicine & Biology. 2006;51(6):1633–48. doi: 10.1088/0031-9155/51/6/019. [DOI] [PubMed] [Google Scholar]
- Hegele RA. Candidate genes, small effects, and the prediction of atherosclerosis. Critical Reviews in Clinical Laboratory Sciences. 1997;34(4):343–67. doi: 10.3109/10408369708998097. [DOI] [PubMed] [Google Scholar]
- Holberg CJ, Halonen M, Solomon S, Graves PE, Baldini M, Erickson RP, Martinez FD. Factor analysis of asthma and atopy traits shows 2 major components, one of which is linked to markers on chromosome 5q. Journal of Allergy & Clinical Immunology. 2001;108(5):772–80. doi: 10.1067/mai.2001.119158. [DOI] [PubMed] [Google Scholar]
- Karasik D, Cupples LA, Hannan MT, Kiel DP. Genome screen for a combined bone phenotype using principal component analysis: the Framingham study. Bone. 2004;34(3):547–56. doi: 10.1016/j.bone.2003.11.017. [DOI] [PubMed] [Google Scholar]
- Klein RF, Turner RJ, Skinner LD, Vartanian KA, Serang M, Carlos AS, Shea M, Belknap JK, Orwoll ES. Mapping quantitative trait loci that influence femoral cross-sectional area in mice. Journal of Bone & Mineral Research. 2002;17(10):1752–60. doi: 10.1359/jbmr.2002.17.10.1752. [DOI] [PubMed] [Google Scholar]
- Lee WT, Cheung AY, Lau J, Lee SK, Qin L, Cheng JC. Bone densitometry: which skeletal sites are best predicted by bone mass determinants? Journal of Bone & Mineral Metabolism. 2004;22(5):447–55. doi: 10.1007/s00774-004-0506-2. [DOI] [PubMed] [Google Scholar]
- Li X, Masinde G, Gu W, Wergedal J, Mohan S, Baylink DJ. Genetic dissection of femur breaking strength in a large population (MRL/MpJ x SJL/J) of F2 Mice: single QTL effects, epistasis, and pleiotropy. Genomics. 2002;79(5):734–40. doi: 10.1006/geno.2002.6760. [DOI] [PubMed] [Google Scholar]
- Li X, Quinones MJ, Wang D, Bulnes-Enriquez I, Jimenez X, De La Rosa R, Aurea GL, Taylor KD, Hsueh WA, Rotter JI, et al. Genetic effects on obesity assessed by bivariate genome scan: the Mexican-American coronary artery disease study. Obesity. 2006;14(7):1192–200. doi: 10.1038/oby.2006.136. [DOI] [PubMed] [Google Scholar]
- Lin HF, Boden-Albala B, Juo SH, Park N, Rundek T, Sacco RL. Heritabilities of the metabolic syndrome and its components in the Northern Manhattan Family Study. Diabetologia. 2005;48(10):2006–12. doi: 10.1007/s00125-005-1892-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livshits G, Deng HW, Nguyen TV, Yakovenko K, Recker RR, Eisman JA. Genetics of bone mineral density: evidence for a major pleiotropic effect from an intercontinental study. Journal of Bone & Mineral Research. 2004;19(6):914–23. doi: 10.1359/JBMR.040132. [DOI] [PubMed] [Google Scholar]
- Martin LJ, Cianflone K, Zakarian R, Nagrani G, Almasy L, Rainwater DL, Cole S, Hixson JE, MacCluer JW, Blangero J, et al. Bivariate linkage between acylation-stimulating protein and BMI and high-density lipoproteins. Obesity Research. 2004;12(4):669–78. doi: 10.1038/oby.2004.77. [DOI] [PubMed] [Google Scholar]
- Masinde GL, Wergedal J, Davidson H, Mohan S, Li R, Li X, Baylink DJ. Quantitative trait loci for periosteal circumference (PC): identification of single loci and epistatic effects in F2 MRL/SJL mice. Bone. 2003;32(5):554–60. doi: 10.1016/s8756-3282(03)00063-2. [DOI] [PubMed] [Google Scholar]
- Mitchell BD, Kammerer CM, Mahaney MC, Blangero J, Comuzzie AG, Atwood LD, Haffner SM, Stern MP, MacCluer JW. Genetic analysis of the IRS. Pleiotropic effects of genes influencing insulin levels on lipoprotein and obesity measures. Arteriosclerosis, Thrombosis & Vascular Biology. 1996;16(2):281–8. doi: 10.1161/01.atv.16.2.281. [DOI] [PubMed] [Google Scholar]
- Musani SK, Huang-Ge Z, Hsu H-C, Yi N-J, Gorman BS, Allison DB. Principal component analysis of quantitative trait loci for immune response to adenovirus in mice. Hereditas. 2006;143:189–197. doi: 10.1111/j.2006.0018-0661.01925.x. [DOI] [PubMed] [Google Scholar]
- Peacock M, Koller DL, Hui S, Johnston CC, Foroud T, Econs MJ. Peak bone mineral density at the hip is linked to chromosomes 14q and 15q. Osteoporosis International. 2004;15(6):489–96. doi: 10.1007/s00198-003-1560-7. [DOI] [PubMed] [Google Scholar]
- R-Development-Core-Team. R: A language and environment for statistical computing. Vienna, Austria: 2007. [Google Scholar]
- Wang X, Kammerer CM, Wheeler VW, Patrick AL, Bunker CH, Zmuda JM. Genetic and environmental determinants of volumetric and areal BMD in multi-generational families of African ancestry: the Tobago Family Health Study. Journal of Bone & Mineral Research. 2007a;22(4):527–36. doi: 10.1359/jbmr.070106. [DOI] [PubMed] [Google Scholar]
- Wang X, Kammerer CM, Wheeler VW, Patrick AL, Bunker CH, Zmuda JM. Pleiotropy and heterogeneity in the expression of bone strength-related phenotypes in extended pedigrees. Journal of Bone & Mineral Research. 2007b;22(11):1766–72. doi: 10.1359/jbmr.070718. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.