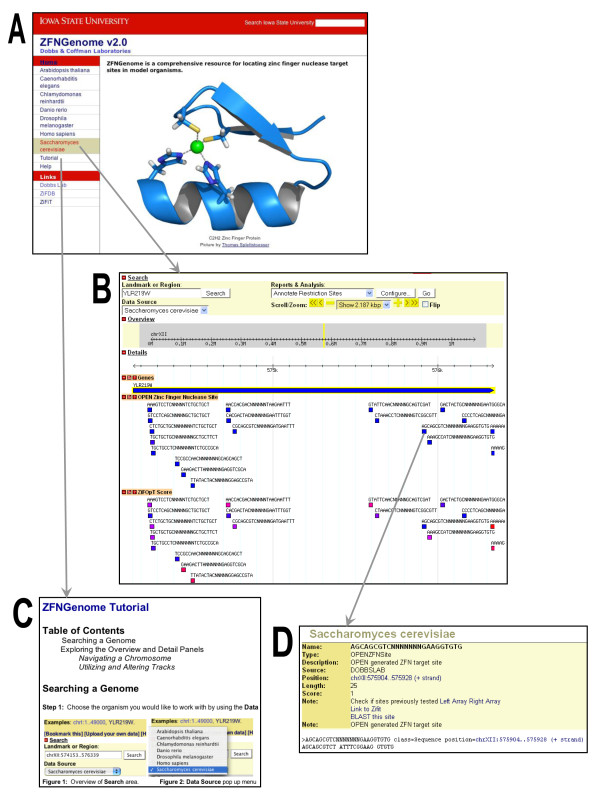
Figure 3.
Examples of resources available in ZFNGenome. (A) The ZFNGenome Homepage is shown. From here, the user can select a model organism from the seven shown in the left hand column. In addition, links to the ZFNGenome Tutorial and Help pages are provided. (B) A screenshot of the result of a search of the S. cerevisiae gene YLR219W is displayed. Key areas of the browser include the search box and the "Scroll/Zoom" areas at the top. The "Overview" and "Detail" panels serve as controls for visualizing the genome. This search shows the single coding region of this gene has 17 potential ZFN target sites, color-coding according to their "uniqueness" and "ZiFOpT" scores (see text). Additional information on each of the tracks can be obtained by clicking on details of the track. For example, clicking on one of the OPEN Zinc Finger Nuclease Sites links the user to details about that specific ZFN target. (C) The ZFNGenome Tutorial offers instructions on navigating the database. The Tutorial can be accessed from the Homepage or from any GBrowse page within ZFNGenome. Help and Instruction links are provided from the GBrowse pages. (D) Clicking on a ZFN target site opens a new window that provides links to ZiFDB, which provides additional information for each zinc finger array, ZiFiT the zinc finger design software that includes the OPEN design method and zinc finger pools, and the BLAST server at National Center for Biotechnology Information (NCBI).