Figure 7.
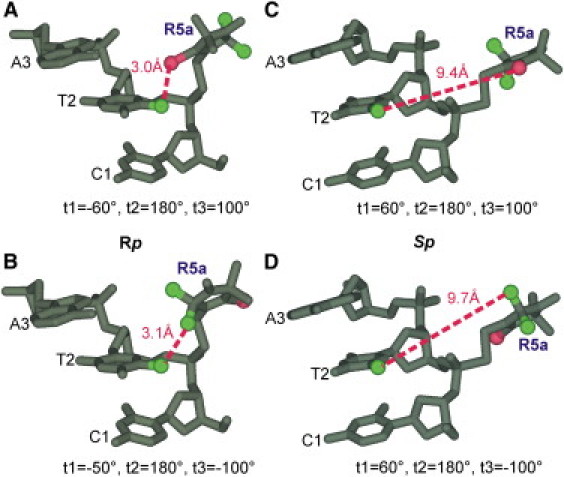
Spatial orientations of Rp and Sp nitroxides at CS2. Molecular models were generated based on the NMR structure using WebLab ViewerPro 3.7 (Molecular Simulations, Orange, CA). During modeling, torsion angles t2 and t3 were set to fixed values (t2 = 180° and t3 = +100° or −100°) based on MD simulations (data not shown), whereas t1 was varied to avoid any steric collisions between R5a and DNA. For Rp-R5a (A and B), the particular rotamers shown give the closest distance (dotted line) between C5-methyl of T2 and functional groups of the R5a pyrroline ring, C4-Br (A) or C2-methyls (B). For Sp-R5a (C and D), none of these pyrroline moieties can be positioned closer than 9 Å to the C5-methyl of T2, and the rotamers shown have t1 arbitrarily set to 60°.
