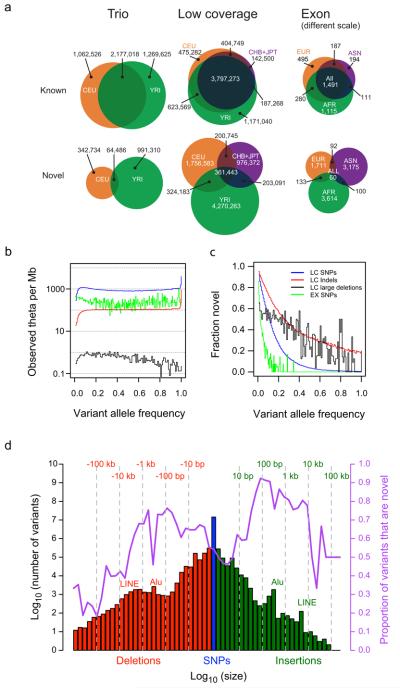
Figure 1. Properties of the variants found.
a, Venn diagrams showing the numbers of SNPs identified in each pilot project in each population or analysis panel, subdivided according to whether the SNP was present in dbSNP release 129 (“Known”) or not (“Novel”). Exon analysis panel AFR is YRI+LWK, ASN is CHB+CHD+JPT, and EUR is CEU+TSI. Note that the scale for the exon project column is much larger than for the other pilots. b, The number of variants per Mb at different allele frequencies divided by the expectation under the neutral coalescent (1/i, where i is the variant allele count), thus giving an estimate of theta per megabase. Blue: low coverage SNPs, red: low coverage indels, black: low coverage genotyped large deletions, green: exon SNPs. The spikes at the right ends of the lines correspond to excess variants for which all samples differed from the reference (approximately 1 per 30 kb), consistent with errors in the reference sequence. c, Fraction of variants in each allele frequency class that were novel. Novelty was determined by comparison to dbSNP release 129 for SNPs and small indels, dbVar (June 2010) for deletions, and two published genomes10, 11 for larger indels. d, Size distribution and novelty of variants discovered in the low coverage project. SNPs are shown in blue, deletions with respect to the reference sequence in red, and insertions or duplications with respect to the reference in green. The fraction of variants in each size bin that were novel is shown by the purple line, and is defined relative to dbSNP (SNPs and indels), dbVar (deletions, duplications, mobile element insertions), dbRIP and other studies49 (mobile element insertions), Venter and Watson genomes10, 11 (indels and deletions), and indels from split capillary reads50 (indels and deletions). To account for ambiguous placement of many indels, discovered indels were deemed to match known indels if they were within 25 bp of a known indel of the same size. To account for imprecise knowledge of the location of most deletions and duplications, discovered variants were deemed to match known variants if they had > 50% reciprocal overlap.