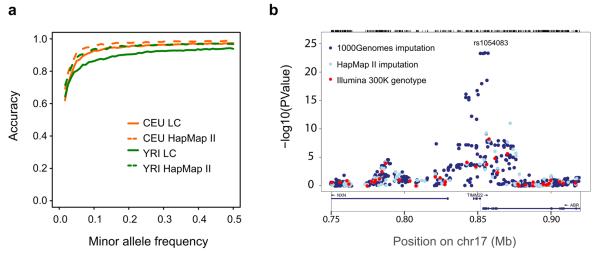
Figure 4. Imputation from the low coverage data.
a, Accuracy of imputing variant genotypes using HapMap 3 sites to impute sites from the low coverage (LC) project into the trio fathers as a function of allele frequency. Accuracy of imputing genotypes from the HapMap II reference panels4 is also shown. Imputation accuracy for common variants was generally a few percent worse from the low coverage project than from HapMap, although error rates increase for less common variants. b, An example of imputation in a cis-eQTL for TIMM22, for which the original Ilumina 300K genotype data gave a weak signal30. Imputation using HapMap data made a small improvement, and imputation using low coverage haplotypes provided a much stronger signal.