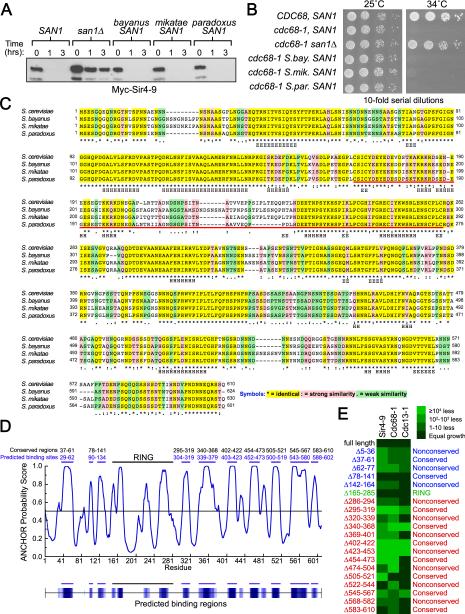
Figure 3. N- & C-terminal regions of San1 are involved in substrate interactions.
(A) Cycloheximide-chase assays of cells expressing Myc-Sir4-9 and San1 homologs were performed to assess complementation. Western blots were probed with anti-Myc antibodies. Time after cycloheximide addition is indicated above each lane. (B) Growth assays of cdc68-1 cells were performed to assess complementation of San1 homologs. Cells were incubated at permissive (25°C) and restrictive temperatures (34°C). (C) ClustalW alignment of S. cerevisiae San1 with its homologs from related Saccharomyces species was performed using Fungal Alignment viewer in the Yeast Genome Database (http://www.yeastgenome.org/cgibin/FUNGI/showAlign). The red bar underlines the RING domain. Secondary structure predicted by Jpred (http://www.compbio.dundee.ac.uk/www-jpred/) is indicated below each residue, with “H” indicating α-helix and “E” indicating β-sheet. (D) ANCHOR prediction of binding sites (http://anchor.enzim.hu/). Blue bars at the top mark the predicted binding sites. Black bars marked conserved regions. (E) Cells expressing the indicated San1 deletion (top) and GAD fusion (left) were spotted media plus or minus histidine to measure spotting efficiency and the 2-hybrid interaction.