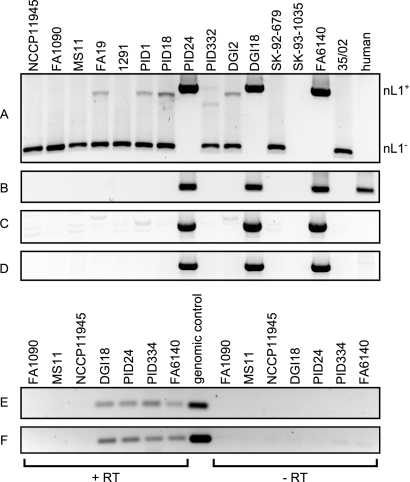
FIG 2 .
PCR amplification of the nL1 fragment in N. gonorrhoeae isolates and nL1 transcript detection by RT-PCR. Purified genomic DNA from sequenced gonococcal isolates and human chromosomal DNA were used as templates for PCR amplification of the nL1 fragment and L1. (A) Primers IS1106for and IRGrev anneal to sequences flanking the nL1 insertion site and yield products of 1,090 and 405 bp for nL1-positive and nL1-negative alleles, respectively. Neither product was detected in DNA from strain SK-93-1035, but this isolate is not predicted to harbor the nL1 fragment. (B) Reactions using primers LINEfor and LINErev, which anneal to sequences internal to the human L1 element and the nL1 fragment. Combinations of flanking and internal primers LINErev and IS1106for (C) or LINEfor and IRGrev (D) were used to confirm the genomic location of nL1. The background bands visible in panels A and C are not due to nL1 sequence amplification, since all of the strains shown here, except PID24, DGI18, and FA6140, were also determined to be nL1 negative by DNA hybridization. Total RNA from nL1-containing isolates (DGI18, PID24, PID334, and FA6140) was used as the template in RT reaction mixtures containing (+RT) or lacking (−RT) reverse transcriptase. The resulting cDNA was amplified with internal nL1 primers L1for and L1rev. Isolates FA1090, MS11, and NCCP11945 lack the L1 fragment and were included as negative controls. Purified genomic DNA from isolate PID334 was used as a positive control. Products were obtained with cDNA generated from positive-strand (E) and negative-strand (F) RNA transcripts.