Abstract
A group of amphiphilic cationic polymers, methoxy polyethylene glycol-block-(polycaprolactone-graft-poly(2-(dimethylamino)ethyl methacrylate)) (PECD), were synthesized by combining ring-opening polymerization (ROP) and atom transfer radical polymerization (ATRP) methods to form nanoparticles (NPs). The structures of these amphiphilic cationic polymers were characterized by 1H NMR measurement. The PECD NPs have hydrophobic cores covered with hydrophilic PEG and cationic PDMAEMA chains. These self-assembly nanoparticles were characterized by dynamic light scattering (DLS) technique. PECD NPs can effectively condense DNA to form compact complexes of the size 65–160 nm suitable for gene delivery. The in vitro gene transfection studies of HeLa and HepG2 cells show that PECD NPs have better transfection efficiency compared to polyethylenimine (PEI) and Lipofectamine 2000 at low dose (N/P = 5). The cytotoxicity result shows that PECD NPs/DNA complexes at the optimal N/P ratio for transfection have comparable toxicity with PEI and Lipofectamine. These results indicate that PECD NPs have a great potential to be used as efficient polymeric carriers for gene transfection.
Keywords: Poly(ε-caprolactone), Gene therapy, Degradable polymers, Poly(2-(dimethylamino ethyl)methacrylate), Amphiphilic copolymers
1. Introduction
Gene therapy has great potentials to treat various genetic diseases. A key hurdle to the clinical applications of gene therapy is lack of safe and effective delivery carriers. Synthetic non-viral delivery carriers, which are safer to use and easier to produce compared to engineered viruses, have increasingly drawn great interests. Cationic polymers are the major types of the non-viral carriers for gene therapy investigated in the past decade. A large number of polycations have been reported to be capable of affecting gene transfection, including branched or linear polyethylenimine (PEI) and its derivatives [1-3], poly (l-lysine) (PLL) [4], polyamidoamine(PAMAM) [5,6], Poly (β-amino ester)s (PAEs) [7-11] and poly(2-(N,N-dimethylamino)ethyl methacrylate) (PDMAEMA) [12-14] etc. These cationic polymers demonstrated attractive features, however, the delivery efficiency still needs to be improved for more effective gene therapy.
Recently, construction of several amphiphilic cationic polymers has been reported [15-23]. Incorporation of hydrophobic components into the cationic polymer backbone is utilized to mimic cationic lipids, which is one of the best transfection reagents and has been used in vivo to treat liver disease as an example. The gene transfection efficiency can be greatly enhanced by hydrophobic modification, which improves cellular uptake through hydrophobic interactions with the cell membranes, and increases endosomal escape of payloads [15,24,25]. These amphiphilic cationic polymers present advantages over lipids, such as capability of versatile modification and without adverse inflammatory responses in vivo [26]. In addition, amphiphilic cationic polymers can also be used to carry hydrophobic anti-cancer drugs. It presents great potential to employ amphiphilic cationic polymers as gene delivery vehicles for cancer treatment in clinic.
Our previous research has shown that PEGylation of PDMAEMA can reduce the cytotoxicity comparing to non-PEGylated homo-polymer PDMAEMA [5]. PEGylated PDMAEMA for DNA vaccine could improve the priming effect and thereby increases the immunogenicity of intranasal administered DNA vaccine. However, this modification strategy has also shown reduction of gene transfection efficiency in vitro [5]. In this study, we have redesigned an amphiphilic cationic polymer, PECD, to enhance gene transfection efficiency. The PECD was prepared by ring-opening polymerization (ROP) and atom transfer radical polymerization (ATRP) methods. DNA condensation ability and physiochemical properties of PECD NPs/DNA complexes, including size and zeta potential, were characterized. In vitro gene transfection efficiency and cytotoxicity were evaluated in HeLa, HepG2 and DRG cells. The endosome escape ability and intracellular distribution of PECD NPs/DNA were measured and compared to other known transfection complexes.
2. Materials and methods
2.1. Materials
γ-(2-Bromo-2-methylpropionate)-ε-caprolactone (BMPCL) was synthesized as reported previously [23,27,28]. ε-Caprolactone (Aldrich) was dried over calcium hydride for 48 h at room temperature and distilled under reduced pressure. Methoxyl poly (ethylene glycol) (Aldrich, Mn = 2000) was dried under vacuum for 24 h at 40 °C. Stannous octanoate, and N,N-dimethylaminoethyl methacrylate (DMAEMA), copper(I) bromide and 2,2′-bipyridine were purchased from Aldrich and used as received. Lipofectamine 2000, Dulbecco’s Modified Eagle’s Medium (DMEM), fetal bovine serum (FBS), penicillin-streptomycin and trypsin were purchased from Invitrogen Corp (Rockville, MD, US). All other reagents were analytical grade and used without further purification.
2.2. Synthesis of macromonomers
Macromonomer, Methoxyl Poly(ethylene glycol)-Poly(caprolactone-co-BMPCL) (mPEG-P(CL-co-BMPCL)), was synthesized by ring-opening polymerization of ε-caprolactone and BMPCL in the presence of mPEG as an initiator and stannous octoate as a catalyst. Briefly, mPEG (Mn = 2000) (1.0 g, 5.0 mmol) was dissolved in 5 mL toluene, and the toluene was distilled off completely to remove the residual water in the reaction system. Then, ε-caprolactone (16.9 g, 148.1 mmol), BMPCL (7.6 g, 74.5 mmol) and stannous octoate (32 μL, 0.08 mmol) were added to the reaction mixtures under dry nitrogen and sealed. The reaction system was stirred at 130 °C for 12 h. Reaction was terminated by cooling the reaction system to room temperature. The polymer was dissolved in methylene chloride and then precipitated by slowly adding cold diethyl ether. The residual solvent was removed under vacuum. The final yields were over 90%.
2.3. Synthesis of PECD
mPEG-b-P(CL-co-BMPCL) (0.2 g, 0.055 mmol) was dissolved in DMAEMA (0.85 g, 5.5 mmol) in schlenk tube. CuBr (8 mg, 0.055 mmol) and 2,2′-bipyridine (16 mg, 0.11 mmol) were added and the mixture was degassed by 3 vacuum/nitrogen cycles. Polymerization was carried out at 60 °C for 12 h. The product was then dissolved in tetrahydrofuran (THF) and recovered by precipitation in cold heptane. Copper catalyst was removed by passing a solution of copolymer in THF through a column of basic alumina. The purified copolymer was recovered by precipitation in cold petroleum ether, filtration and drying under reduced pressure at 40 °C.
2.4. Polymer characterization
The molecular weights of polymers were determined by gel permeation chromatography (GPC) and chemical structure by NMR spectroscopy. The gel permeation chromatography system (GPC, Agilent 1100, Santa Clara, CA) was used to measure the molecular weight and molecular weight distribution of polymers. THF was used as the eluent at a low flow rate of 1.0 mL/min. Monodispersed polystyrene standards were used to generate the calibration curve. 1H NMR spectra were recorded on a Varian INOVA 500 MHz NMR machine, using CDCl3 as solvent and tetramethylsilane (TMS) as internal standard.
2.5. Preparation of nanoparticles (NPs)
NPs were prepared using nanoprecipitation technology. Briefly, 30.0 mg PECD1 (as shown in Table 1) was dissolved in 1 mL THF, which is slowly added to 10 mL double distilled water. The mixture was stirred at room temperature to remove THF, and the final volume was adjusted to 10 mL for further experiments. The pH value of PECD1 solution was adjusted to 7.3 by 1.0 m HCl.
Table 1.
Molecular characterization of graft copolymers
The design number of DMAEMA per graft.
The determined number of DMAEMA per graft by 1H NMR.
The Mn of graft copolymers was estimated by 1H NMR.
2.6. Preparation of NPs/DNA of complexes
For preparation of complexes, DNA plasmid was diluted to 1 μg/50 μL in 10 mm HEPES (pH = 7.3). Complexes were prepared by adding NPs solution to equal volumes of plasmid solution (containing 1 μg DNA) at various N/P ratios by pipetting and incubated for 30 min before characterization and gene transfection experiments.
2.7. Agarose gel electrophoresis
To assess DNA condensation ability of the NPs, agarose gel electrophoresis was performed. The PECD NPs/DNA complexes with different N/P ratios ranging from 0.5 to 10 were prepared as described above. 20 μL of the NPs/DNA (0.2 μg) complexes solution was mixed with 4 μL 6x loading buffer and loaded into a 0.8 wt% agarose gel containing 0.5 μg/mL ethidium bromide. Electrophoresis was set up in 1x TAE buffer at 120 V and kept for 40 min. DNA retardation was analyzed on UV illuminator to indicate the location of the DNA.
2.8. Characterization of particle sizes and zeta potential
The particle sizes and zeta potential of PECD NPs and PECD NPs/DNA complexes were measured using a Zetasizer 3000HS (Malvern Instrument, Inc., Worcestershire, UK) at a wavelength of 677 nm with a constant angle of 90 °. Complexes solutions (200 μL) containing 2 μg of plasmid were prepared at various N/P ratios from 5 to 20 and diluted with 0.8 mL of double distilled water before characterization.
2.9. Luciferase assay
Prior to transfection, HepG2 cells and HeLa cells were seeded in 24 well plate at an initial density of 3 × 104 cells per well in 0.5 mL growth medium and incubated for 18–20 h to achieve 70–80% confluence. The media was replaced by 0.5 mL Opti-MEM containing PECD NPs/pGL3.0 complexes (containing 0.5 μg plasmid) at various N/P ratios and continuously incubated for 4 h. Serum free media was then replaced by complete culture medium and incubated for additional 24 h. After incubation, the medium was removed and the cells were rinsed twice with PBS. 0.2 mL 1x Reporter Lysis Buffer (Promega Co., Madison, WI) was then added to each well to lyse the cells. The cell suspension was frozen in −80 °C for 30 min and then thawed. The cell lysate was transferred into a 1 mL centrifuge vial and centrifuged for 30 s at 12,000 rpm. The supernatant was collected for luminescence measurements. Following the manufacturer’s protocol for luciferase assay (Promega, Madison, WI), relative light units (RLU) were measured with chemiluminometer (Autolumat, LB953, EG&G Berthold, Germany). RLUs were normalized to protein concentrations in the cell extracts measured by Pierce BCA (bicinchoninic acid) Protein Assays (Thermo Fisher Scientific Inc, Rockford, IL).
2.10. Green fluorescent protein assay
Transfection of pEGFP-N1 plasmid mediated by PECD NPs in HeLa cells and HepG2 cells were also evaluated by the method as mentioned above in the luciferase assay section. After additional 24 h incubation, the cells were directly observed by an inverted microscope (Olympus IX 70, Olympus, Tokyo, Japan). The microscopy images were recorded using Cool SNAP-Pro (4.5.1.1) software.
2.11. Cell viability
The cytotoxicity of PECD NPs/DNA complexes was assessed by Cell Counting Kit-8 (CCK-8) (Dojindo Molecular Technologies, Gaithersburg, MD). HeLa and HepG2 cells were seeded in a 96-well plate at a density of 1 × 104 cells/well and subsequently transfected using similar protocol as described before. After 24 h incubation, 10 mL of thawed CCK-8 solution was added to each well. After further incubation for 2 h, the absorbance was measured at 450 nm by Infinite M200 (TECAN, Männedorf, Switzerland). The results were expressed as the mean percentage of cell viability relative to untreated cells.
2.12. Confocal laser scanning microscope (CLSM)
pGL3.0 plasmid was labeled with Cy5 by using the Label IT Nucleic Acid Labeling Kit (Mirus, Madison, WI) according to the manufacturer’s protocol. HepG2 cells were plated in 6-well plate with glass cover slip at bottom at 2 × 105 cells/well one day before transfection. After the culture medium was changed to Opti-MEM, well incubated complexes solution containing 1 μg Cy5-labeled plasmid (N/P = 10) was added into each plate. After 4 h incubation at 37 °C in 5% CO2 humidified atmosphere, the transfection solutions were aspirated and substituted with complete culture medium. After additional 8 h incubation, the cells were washed with PBS for three times before acidic late endosome staining with LysoTracker blue (Molecular Probes, Eugene, OR). Then, the intracellular distribution was observed with a confocal microscopy (Zeiss LSM510, Carl Zeiss Shanghai Co. Ltd, Shanghai, China). The colocalization ratio of Cy5-DNA with late endosomes/lysosomes was calculated as described elsewhere [29]. The results are presented as mean and standard deviation obtained from 7 cells. P value was calculated by Student’s t test, between the result of the cells treated with PEI/Cy5-DNA complexes and that of the cells treated with PECD3 NPs/Cy5-DNA complexes. The symbol * means P < 0.05.
3. Results and discussion
3.1. Synthesis of PECD
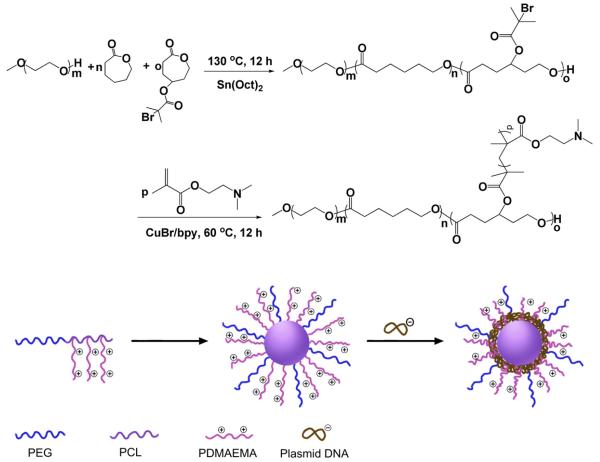
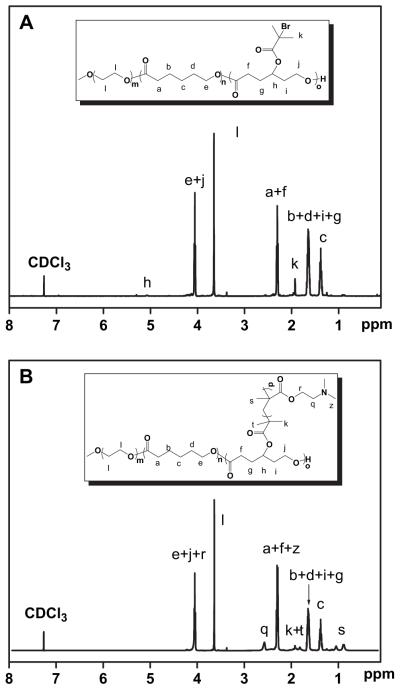
PECD was synthesized by combination of ring-opening polymerization (ROP) and atom transfer radical polymerization (ATRP) technique (shown in Scheme 1). γ-(2-Bromo-2-methylpropionate)-ε-caprolactone (BMPCL) is one of the monomers which can be used to synthesize degradable polyester copolymers bearing ATRP initiating group [27]. Macroinitiator, mPEG-P(CL-co-BMPCL), was readily prepared by ring-opening polymerization of ε-caprolactone and BMPCL using mPEG (Mn = 2000) as initiator and Sn(Oct)2 as a catalyst. As shown in Fig .1A, the characteristic peaks of PCL moieties at 1.36 ppm and BMPCL moieties at 1.92 ppm in 1H NMR spectrum are clearly displayed, and used to calculate the composition and molecular weight of mPEG-P(CL-co-BMPCL) by comparing the intensities of these two signals to that of mPEG at 3.65 ppm. Additionally, no characteristic peak arising from residual BMPCL monomer at 2.47–3.02 ppm was found, indicating the mPEG-P(CL-co-BMPCL) copolymer was successfully obtained. The Mn of mPEG-P (CL-co-BMPCL) determined by 1H NMR is 1.50 × 104 g/mol, and the mean number of BMPCL in the copolymer is 4.0.
Scheme 1.
Synthesis route of PECD and preparation of PECD NPs with plasmid DNA as payload.
Fig. 1.
1H NMR spectra of mPEG-b-P(CL-co-BMPCL) (A) and PECD1 (B).
Then, the ATRP polymerization of DMAEMA was performed in bulk at 60 °C for 12 h. The results of polymerization are shown in Table 1. The chemical structure of PECD was further confirmed by 1H NMR using CDCl3 as solvent. Characteristic peak of vinyl protons is no longer visible between 5 and 6 ppm, which is an indication of the absence of residual DMAEMA monomers (Fig. 1B). Fig. 1B presents the detailed assignments of 1H NMR spectrum of PECD1. Compared to Fig. 1A, there appears some new peaks belonging to DMAEM (δ = 2.55 ppm). Taking these into account, PECDs were successfully synthesized. The Mn values of PDMAEMA were estimated by comparing the intensities of signals at 2.55 ppm and 3.65 ppm which belong to PDMAEMA moieties and mPEG moieties, respectively. Although, the ATRP polymerization was performed in bulk, it is noted that the results showed that the determined polymerization degree of DMAEMA per graft was well correlated with the design polymerization degree (Table 1). Hence, PECDs were successfully synthesized.
3.2. Characterization of PECD NPs and its complexes with DNA
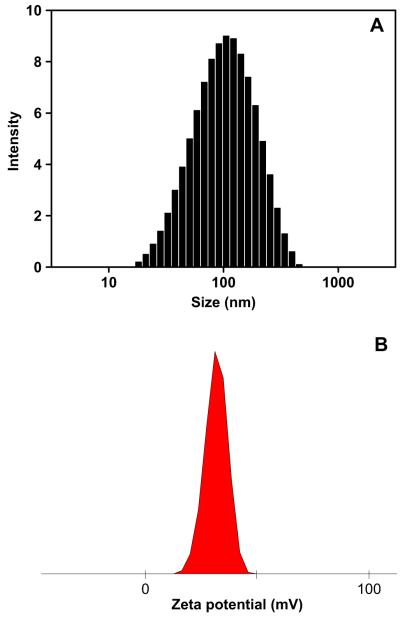
PECD was one of amphiphilic copolymers, which can be assembled into core-shell structure nanoparticles in aqueous solution. PECD NPs were prepared by nanoprecipitation technique. DLS was performed to measure the hydrodynamic diameter of PECD NPs and their complexes with DNA. The mean sizes of nanoparticles formed by PECD1, PECD2 and PECD3 were 56.3 ± 0.5 nm, 77.4 ± 0.2 nm and 85.2 ± 1.6 nm (Fig. 2A) in diameter, respectively. The positive charged surface of PECD NPs was also confirmed by zeta potential measurements. Zeta potentials of PECD1 NPs, PECD2 NPs and PECD3 NPs were 28.9 ± 0.4 mv, 32.0 ± 0.5 mv and 31.5 ± 0.6 mv (Fig. 2B).
Fig. 2.
Hydrodynamic diameter (A) and zeta potential (B) of PECD3 NPs.
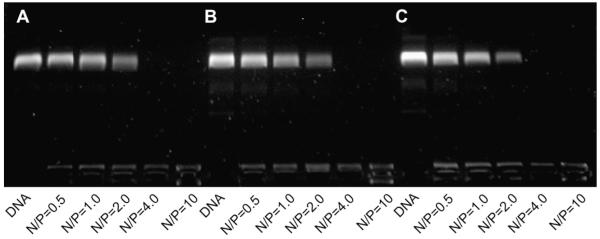
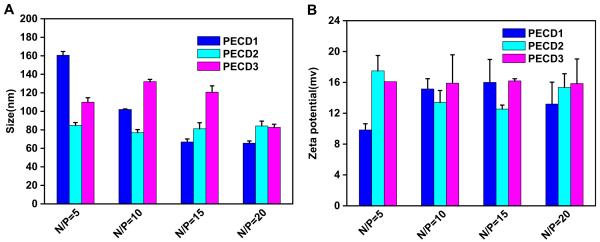
To investigate the condensation capability of PECD NPs with DNA, agarose gel electrophoresis was performed at N/P ratios ranging from 0 to 10. As shown in Fig. 3, the mobility of plasmid was completely retarded at the ratio higher than 2, indicating that all copolymers could bind DNA strongly. And there is no significant difference in DNA binding ability for all the copolymers. The formation of polymer-DNA complexes with appropriate size and positive charged surface is an important prerequisite for polycations used as gene carriers to be internalized into cells. To investigate the interaction between DNA and cationic PECD NPs, dynamic light scattering was performed to measure the particle sizes and zeta potential. As shown in Fig. 4A, all the polymers could condense DNA into nanosized complexes with the size range of 65–160 nm, which is suitable to be applied for endocytotic cellular uptake [30]. Zeta potential of the complexes at various N/P ratios was shown in Fig. 4B. All complexes presented net positive charges. For the complexes with the N/P ratio higher than 10, the zeta potential approached to a plateau due to saturation of polycations complexed with DNA [31].
Fig. 3.
Agarose gel electrophoresis retardation assay of the copolymers at various N/P ratios of PECD NPs/DNA complexes. (A) PECD1 NPs, (B) PECD2 NPs and (C) PECD3 NPs.
Fig. 4.
Zeta potential and particle size measurement of PECD NPs/DNA complexes at various N/P ratios.
3.3. In vitro transfection
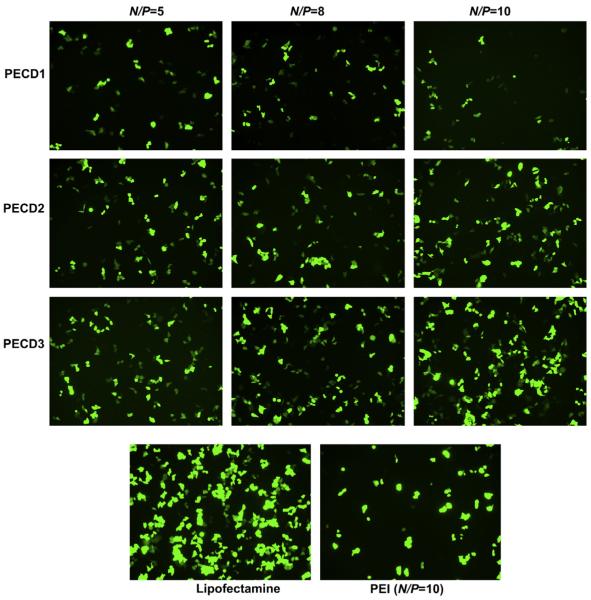
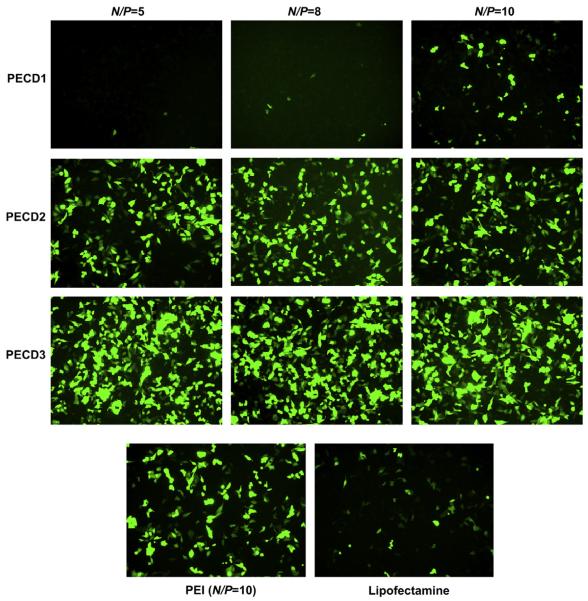
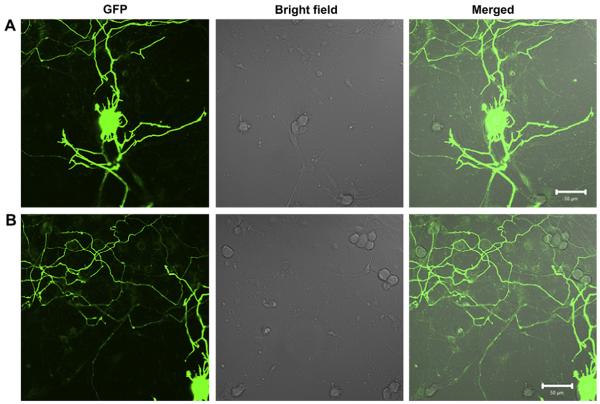
To investigate gene transfection efficiency of PECD NPs in vitro, HeLa and HepG2 cells were transfected with PECD NPs/EGFP-N1 or pGL3.0 complexes. Fig. 5 shows the green fluorescence images of the transfected HeLa cells at N/P ratios from 5 to 10 for PECD NPs. PECD3 NPs with the longest cationic block has relatively higher gene transfection capability, whereas the gene transfection capability of PECD1 and PECD2 is lower. The optimal transfection of PECD3 NPs at N/P ratio of 10 was comparable to PEI (N/P = 10), it is notable that the transfection efficiency is not significantly affected by varying N/P ratio for PECD NPs. This is also confirmed in the HepG2 cells study (Fig. 6). As shown in Fig. 6, the transfection results were in accordance with that obtained in HeLa cells. The transfection efficiency was nearly the same to each individual PECD NPs with different N/P ratios. The PECD2 and PECD3 NPs present much better transfection efficiency than that of commercial gold standard, Lipofectamine and PEI. The PECD2 and PECD3 NPs can transfer EGFP plasmid in HepG2 cells effectively even at N/P ratio of 5, which indicates that the amount of transfection reagent is significantly reduced, compared to other polycations carriers. These results demonstrate that the gene transfer capability of PECD NPs is dependent on cell line and also depends on N/P ratios. For many neuron cells types such as primary dorsal root ganglion (DRG) cells, transfection efficiencies obtained by conventional methods are usually very low. It is a crucial prerequisite to transfect with high efficiencies for successful biomolecular experiments in neuroscience research. Here, it is surprising that PECD delivery system also shows good transfection efficiency and maintains intact neuronal morphology on DRG neuron cells (As shown in Fig. 7). This result indicates that PECD system may be very useful to be employed for the biological function research of neuron cells.
Fig. 5.
Fluorescence images of HeLa cells transfected by PECD NPs/EGFP-N1 plasmid complexes at N/P ratios from 5 to 10. Lipofectamine 2000 and PEI (at its optimal N/P of 10) were used as positive controls. Scale bar is 30 μm.
Fig. 6.
Fluorescence images of HepG2 cells transfected by PECD NPs/EGFP-N1 plasmid complexes at N/P ratios from 5 to 10. Lipofectamine 2000 and PEI (at its optimal N/P of 10) were used as positive controls. Scale bar is 30 μm.
Fig. 7.
Confocal fluorescence images of DRG cells transfected by PECD3 NPs/EGFP-N1 plasmid complexes (using 1.0 μg plasmid) at N/P ratios from 10. Both A and B are representative different parts of same cells. Scale bar was 50 μm.
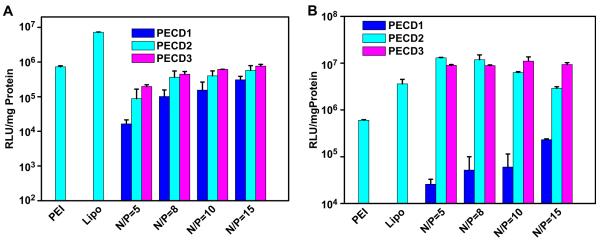
The in vitro transfection efficiency of PECD NPs/DNA complexes was also evaluated using luciferase as a reporter gene in HeLa and HepG2 cells. Fig. 8 shows the gene transfection efficiency of PECD NPs for DNA delivery in comparison with those of branched PEI (25 kDa) and Lipofectamine 2000, which are the best commonly used transfection reagents. It was found that gene transfection efficiency was greatly dependent on the cell type, molecular weight of cationic PDMAEMA grafts and N/P ratio of the PECD NPs/DNA complexes. Similar to GFP transfection results, PECD NPs were more effective in transfecting HepG2 cells than that in HeLa cells. In HepG2 cells, the optimal N/P ratio of PECD2 and PECD3 NPs was around 5, which was lower than N/P = 10 of PEI. The transfection efficiency was reduced with increased N/P ratios. Furthermore, transfection efficiency of PECD2 and PECD3 NPs at N/P of 5 was much better than that of PEI and Lipofectamine 2000 in HepG2 cells. However, PECD1 NPs presents lower transfection level, probably due to low molecular weight of cationic block, it is also previously reported in literature [20,30]. In contrast, the transfection efficiency was enhanced with increased N/P ratios in HeLa cells. The optimal N/P ratio of PECD2 and PECD3 was 15 and transfection efficiency of PECD2 and PECD3 NPs at N/P in the range from 5 to 15 was comparable to that of PEI.
Fig. 8.
In vitro transfection efficiency of PECD NPs/pGL3.0 complexes in HeLa (A) and HepG2 (B) cell lines at various N/P ratios. Transfection was performed at a dose of 0.5 μg of DNA (mean ± SD n = 3).
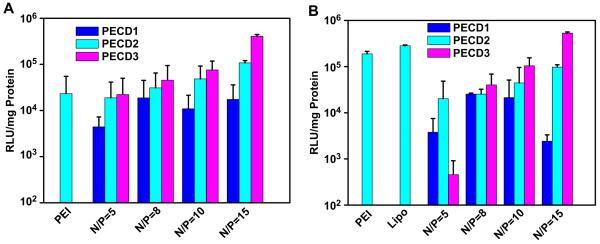
Luciferase expression levels for PECD2 and PECD3 NPs were higher than those of PEI 25K in HeLa and HepG2 cell lines, respectively. Similar results that hydrophobicity enhanced the transfection efficiency of carriers have also been reported by other groups[17,18,20,24,30,32,33]. This is believed that the presence of hydrophobic segments in carriers improves cellular uptake through hydrophobic interactions with the cell membranes and thereby enhances endosomal escape. For practical applications, the gene transfection occurs in the presence of serum, which usually reduces the transfection efficiency of cationic carriers. To verify this hypothesis, a transfection experiment using pGL3.0 plasmid was also performed in the presence of 10% serum. As shown in Fig. 9, the level of gene expression was enhanced for transfection of PECD2 and PECD3 NPs with increased N/P ratio. Both PECD2 and PECD3 NPs presented the highest efficiency at N/P ratio of 15 in HeLa and HepG2 cells. Notably, the transfection efficiency of PECD3 NPs at N/P ratio of 15 was higher than that of PEI and Lipofectamine 2000. These results show that PECD NPs delivery system has potential to be used as an efficient gene delivery system in vivo.
Fig. 9.
In vitro transfection efficiency of PECD NPs/pGL3.0 complexes in HeLa (A) and HepG2 (B) cell lines at various N/P ratios at the presence of 10% FBS. Transfection was performed at a dose of 0.5 μg of DNA (mean ± SD n = 3).
3.4. Cytotoxicity of PECD NPs/DNA complexes
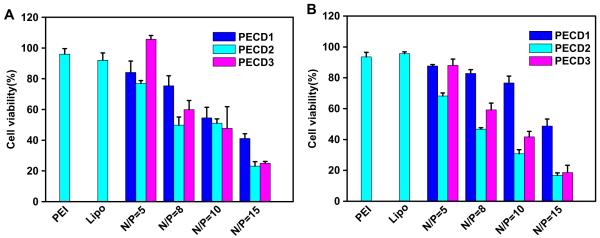
The CCK-8 assay was performed to evaluate the in vitro cytotoxicity of PECD NPs/DNA complexes (as shown in Fig. 10). In both cell lines, cytotoxicity of PECD NPs/DNA complexes was greatly dependent on molecular weight of PDMAEMA grafts and N/P ratios. It was found that all PECDs demonstrated comparable cytotoxicity to PEI and Lipofectamine at their optimal N/P ratio of 5. However, PECD NPs/DNA complexes present high cytotoxicity to cells at high N/P ratios. As discussed above, the incorporation of hydrophobic block into the carriers would enhance the interactions between complexes and cell membranes when it infused with plasma membrane for penetrating into cytoplasm. However, cell membrane was greatly damaged at higher N/P ratios.
Fig. 10.
Cell viabilities of PECD NPs/DNA complexes at different N/P ratios in vitro were evaluated using HeLa (A) and HepG2 (B) cells determined by CCK-8 assay.
3.5. Intracellular distribution of PECD NPs/DNA binary complexes
It is known that the successful escape of gene carriers from endosomes is crucial to improve gene transfection efficiency. Confocal microscopy was used to observe the intracellular distribution of PECD3 NPs/DNA complexes at N/P ratio of 10. pGL3.0 plasmid labeled by Cy5 dye and endosomes stained with Lyso-Tracker green were employed to identify the localization of PECD3 NPs/DNA complexes. Cell nuclei were stained by Hoechst 33342 in live cells. Accordingly, pGL3.0 plasmid that co-localized with late endosomes/lysosomes should be observed as yellow. For the transfection experiment, cells were incubated with PECD3 NPs/DNA complexes for 4 h and CLSM images were taken after further incubation for 8 h.
Fewer obvious yellow regions were observed in Fig. 11C compared to Fig. 11B, it indicated that more PECD3 NPs/DNA complexes were visible in the cells and released into cytoplasm while not in the endosome. Based on the numbers of red and yellow pixels in the CLSM images, the colocalization ratio of Cy5-DNA with late endosomes/lysosomes was further estimated. As shown in Fig. 11D, the colocalization ratio was approximately 61% for the cells treated with PEI/DNA complexes, whereas that for the cells treated with PECD3 NPs/DNA complexes was around 36%, it demonstrated that PECD3 NPs/DNA complexes were more effective to escape from late endosomes/lysosomes. PEI has been known to be an effective transfection reagent due to “proton sponge” effect, which facilitates the escape of complexes from endosomes. Therefore, these results suggest that amphiphilicity may contribute to facilitate endosomal escape of the PECD NPs/DNA complexes and enhance transfection efficiency.
Fig. 11.
In vitro confocal images of negative control (A), PEI/DNA complexes at N/P ratio of 10 (B) and PECD3 NPs/DNA complexes at N/P ratio of 10 (C). Confocal images of HepG2 cells transfected with PECD3 NPs/DNA complexes were taken after transfection for 12 h. The late endosome and lysosome network were stained with LysoTracker green. Plasmid DNA was labeled with Cy5 and cell nuclei were stained by Hoechst 33342. Each scale bar represents 20 μm. (D) Colocalization ratio of Cy5-labeled DNA with LysoTracker (late endosomes/lysosomes) calculated by pixel counting (number of counted cells: 7, *p < 0.05).
4. Conclusions
A series of amphiphilic PECD were synthesized via ROP and ATRP polymerization methods. At N/P ratio of 2 and above, all PECDs NPs can effectively bind plasmid DNA to form complexes with sizes around 65–160 nm and positive zeta potentials about 10–18 mv. In vitro gene transfection efficiency depends on the type of cell lines, the molecular weight of PDMAEMA grafts and N/P ratios of carriers. PECDs NPs show much better transfection efficiency than Lipofectamine 2000 and PEI in HepG2 cells and comparable efficiency to PEI in HeLa cells. The increase in the length of PDMAEMA moiety improves transfection efficiency but decreases cell viabilities. In addition, confocal images indicate that PECD NPs/DNA complexes have strong escape ability out of endosomes.
Acknowledgements
We thank Professor Zhuan Zhou (Institute of Molecular Medicine, PekingUniversity) for providing DRG cells. This project was supported by a grant from National Key Basic Research Program of China (2009CB930200), National Grand Program on Key Infectious Disease Control (2008ZX10001-015-10), National Natural Science Foundation of China (No.30970784 and 30772007), Tianjin Natural Science Foundation (No. 09JCYBJC13800), NIH/NCRR/RCMI 2 G12 RR003048, Specialized Research Fund for the Doctoral Program of Higher Education (20090032120013). Authors declare there are no any conflicts of interest that relate to papers accepted for publication.
Appendix
Figure with essential color discrimination. Figs. 2,4-11 in this article is difficult to interpret in black and white. The full color images can be found in the on-line version, at doi:10.1016/j.biomaterials.2010.09.052.
References
- [1].Boussif O, Lezoualch F, Zanta MA, Mergny MD, Scherman D, Demeneix B, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in-vivo - polyethylenimine. P Natl Acad Sci USA. 1995;92:7297–301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kursa M, Walker GF, Roessler V, Ogris M, Roedl W, Kircheis R, et al. Novel shielded transferrin-polyethylene glycol-polyethylenimine/DNA complexes for systemic tumor-targeted gene transfer. Bioconjug Chem. 2003;14:222–31. doi: 10.1021/bc0256087. [DOI] [PubMed] [Google Scholar]
- [3].Tarcha PJ, Pelisek J, Merdan T, Waters J, Cheung K, von Gersdorff K, et al. Synthesis and characterization of chemically condensed oligoethylenimine containing beta-aminopropionamide linkages for sirna delivery. Biomaterials. 2007;28:3731–40. doi: 10.1016/j.biomaterials.2007.04.037. [DOI] [PubMed] [Google Scholar]
- [4].Choi YH, Liu F, Kim JS, Choi YK, Park JS, Kim SW. Polyethylene glycol-grafted poly-l-lysine as polymeric gene carrier. J Control Release. 1998;54:39–48. doi: 10.1016/s0168-3659(97)00174-0. [DOI] [PubMed] [Google Scholar]
- [5].Qiao Y, Huang Y, Qiu C, Yue XY, Deng LD, Wan YM, et al. The use of pegylated poly [2-(n, n-dimethylamino) ethyl methacrylate] as a mucosal DNA delivery vector and the activation of innate immunity and improvement of hiv-1-specific immune responses. Biomaterials. 2010;31:115–23. doi: 10.1016/j.biomaterials.2009.09.032. [DOI] [PubMed] [Google Scholar]
- [6].Bielinska AU, Chen CL, Johnson J, Baker JR. DNA complexing with polyamidoamine dendrimers: implications for transfection. Bioconjug Chem. 1999;10:843–50. doi: 10.1021/bc990036k. [DOI] [PubMed] [Google Scholar]
- [7].Brito L, Little S, Langer R, Amiji M. Poly(beta-amino ester) and cationic phospholipid-based lipopolyplexes for gene delivery and transfection in human aortic endothelial and smooth muscle cells. Biomacromolecules. 2008;9:1179–87. doi: 10.1021/bm7011373. [DOI] [PubMed] [Google Scholar]
- [8].Zugates GT, Anderson DG, Little SR, Lawhorn IEB, Langer R. Synthesis of poly (beta-amino ester)s with thiol-reactive side chains for DNA delivery. J Am Chem Soc. 2006;128:12726–34. doi: 10.1021/ja061570n. [DOI] [PubMed] [Google Scholar]
- [9].Little SR, Lynn DM, Puram SV, Langer R. Formulation and characterization of poly (beta amino ester) microparticles for genetic vaccine delivery. J Control Release. 2005;107:449–62. doi: 10.1016/j.jconrel.2005.04.022. [DOI] [PubMed] [Google Scholar]
- [10].Little SR, Lynn DM, Ge Q, Anderson DG, Puram SV, Chen JZ, et al. Poly-beta amino ester-containing microparticles enhance the activity of nonviral genetic vaccines. P Natl Acad Sci USA. 2004;101:9534–9. doi: 10.1073/pnas.0403549101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Akinc A, Anderson DG, Lynn DM, Langer R. Synthesis of poly(beta-amino ester)s optimized for highly effective gene delivery. Bioconjug Chem. 2003;14:979–88. doi: 10.1021/bc034067y. [DOI] [PubMed] [Google Scholar]
- [12].Layman JM, Ramirez SM, Green MD, Long TE. Influence of polycation molecular weight on poly(2-dimethylaminoethyl methacrylate)-mediated DNA delivery in vitro. Biomacromolecules. 2009;10:1244–52. doi: 10.1021/bm9000124. [DOI] [PubMed] [Google Scholar]
- [13].Lin S, Du F, Wang Y, Ji S, Liang D, Yu L, et al. An acid-labile block copolymer of pdmaema and peg as potential carrier for intelligent gene delivery systems. Biomacromolecules. 2008;9:109–15. doi: 10.1021/bm7008747. [DOI] [PubMed] [Google Scholar]
- [14].Jiang X, Lok MC, Hennink WE. Degradable-brushed phema-pdmaema synthesized via atrp and click chemistry for gene delivery. Bioconjug Chem. 2007;18:2077–84. doi: 10.1021/bc0701186. [DOI] [PubMed] [Google Scholar]
- [15].Sun TM, Du JZ, Yan LF, Mao HQ, Wang J. Self-assembled biodegradable micellar nanoparticles of amphiphilic and cationic block copolymer for sirna delivery. Biomaterials. 2008;29:4348–55. doi: 10.1016/j.biomaterials.2008.07.036. [DOI] [PubMed] [Google Scholar]
- [16].Xiong XB, Uludag H, Lavasanifar A. Biodegradable amphiphilic poly(ethylene oxide)-block-polyesters with grafted polyamines as supramolecular nano-carriers for efficient sirna delivery. Biomaterials. 2009;30:242–53. doi: 10.1016/j.biomaterials.2008.09.025. [DOI] [PubMed] [Google Scholar]
- [17].Zhang K, Fang HF, Wang ZH, Li Z, Taylor JSA, Wooley KL. Structure-activity relationships of cationic shell-crosslinked knedel-like nanoparticles: shell composition and transfection efficiency/cytotoxicity. Biomaterials. 2010;31:1805–13. doi: 10.1016/j.biomaterials.2009.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhang K, Fang HF, Wang ZH, Taylor JSA, Wooley KL. Cationic shell-crosslinked knedel-like nanoparticles for highly efficient gene and oligonucleotide transfection of mammalian cells. Biomaterials. 2009;30:968–77. doi: 10.1016/j.biomaterials.2008.10.057. [DOI] [PubMed] [Google Scholar]
- [19].Zhu CH, Jung S, Luo SB, Meng FH, Zhu XL, Park TG, et al. Co-delivery of sirna and paclitaxel into cancer cells by biodegradable cationic micelles based on pdmaema-pcl-pdmaema triblock copolymers. Biomaterials. 2010;31:2408–16. doi: 10.1016/j.biomaterials.2009.11.077. [DOI] [PubMed] [Google Scholar]
- [20].Zhu JL, Cheng H, Jin Y, Cheng SX, Zhang XZ, Zhuo RX. Novel polycationic micelles for drug delivery and gene transfer. J Mater Chem. 2008;18:4433–41. [Google Scholar]
- [21].Wang Y, Gao SJ, Ye WH, Yoon HS, Yang YY. Co-delivery of drugs and DNA from cationic core-shell nanoparticles self-assembled from a biodegradable copolymer. Nat Mater. 2006;5:791–6. doi: 10.1038/nmat1737. [DOI] [PubMed] [Google Scholar]
- [22].Qiu LY, Bae YH. Self-assembled polyethylenimine-graft-poly(epsilon-caprolactone) micelles as potential dual carriers of genes and anticancer drugs. Biomaterials. 2007;28:4132–42. doi: 10.1016/j.biomaterials.2007.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Guo S, Qiao Y, Wang W, Xing J, Deng L, Dong A, et al. Synthesis and properties of polycaprolactone-graft-poly(2-(dimethylamino)ethyl methacrylate-co-methoxy polyethylene glycol monomethacrylate) as non-viral gene vector. Polym Adv Technol. 2010 doi:10.1002/pat.1694. [Google Scholar]
- [24].Sharma R, Lee JS, Bettencourt RC, Xiao C, Konieczny SF, Won YY. Effects of the incorporation of a hydrophobic middle block into a peg-polycation diblock copolymer on the physicochemical and cell interaction properties of the polymer-DNA complexes. Biomacromolecules. 2008;9:3294–307. doi: 10.1021/bm800876v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Anderson DG, Lynn DM, Langer R. Semi-automated synthesis and screening of a large library of degradable cationic polymers for gene delivery. Angew Chem Int Ed. 2003;42:3153–8. doi: 10.1002/anie.200351244. [DOI] [PubMed] [Google Scholar]
- [26].Blow N. Delivering the future. Nature. 2007;450:1117–22. doi: 10.1038/4501117a. [DOI] [PubMed] [Google Scholar]
- [27].Mecerreyes D, Atthoff B, Boduch KA, Trollsas M, Hedrick JL. Unimolecular combination of an atom transfer radical polymerization initiator and a lactone monomer as a route to new graft copolymers. Macromolecules. 1999;32:5175–82. [Google Scholar]
- [28].Guo S, Wang W, Deng L, Xing J, Dong A. Poly(caprolactone)-graft-poly(2-‘(dimethylamino)ethyl methacrylate) amphiphilic copolymers prepared via a combination of rop and atrp: synthesis, characterization, and self-assembly behavior. Macromol Chem Phys. 2010;211:1572–8. [Google Scholar]
- [29].Takemoto H, Ishii A, Miyata K, Nakanishi M, Oba M, Ishii T, et al. Polyion complex stability and gene silencing efficiency with a sirna-grafted polymer delivery system. Biomaterials. 2010;31:8097–105. doi: 10.1016/j.biomaterials.2010.07.015. [DOI] [PubMed] [Google Scholar]
- [30].Arote R, Kim TH, Kim YK, Hwang SK, Jiang HL, Song HH, et al. A biodegradable poly(ester amine) based on polycaprolactone and polyethylenimine as a gene carrier. Biomaterials. 2007;28:735–44. doi: 10.1016/j.biomaterials.2006.09.028. [DOI] [PubMed] [Google Scholar]
- [31].Lu B, Xu XD, Zhang XZ, Cheng SX, Zhuo RX. Low molecular weight polyethylenimine grafted n-maleated chitosan for gene delivery: properties and in vitro transfection studies. Biomacromolecules. 2008;9:2594–600. doi: 10.1021/bm8004676. [DOI] [PubMed] [Google Scholar]
- [32].Yang Z, Sahay G, Sriadibhatla S, Kabanov AV. Amphiphilic block copolymers enhance cellular uptake and nuclear entry of polyplex-delivered DNA. Bioconjug Chem. 2008;19:1987–94. doi: 10.1021/bc800144a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Gebhart CL, Sriadibhatla S, Vinogradov S, Lemieux P, Alakhov V, Kabanov AV. Design and formulation of polyplexes based on pluronic-polyethyleneimine conjugates for gene transfer. Bioconjug Chem. 2002;13:937–44. doi: 10.1021/bc025504w. [DOI] [PubMed] [Google Scholar]