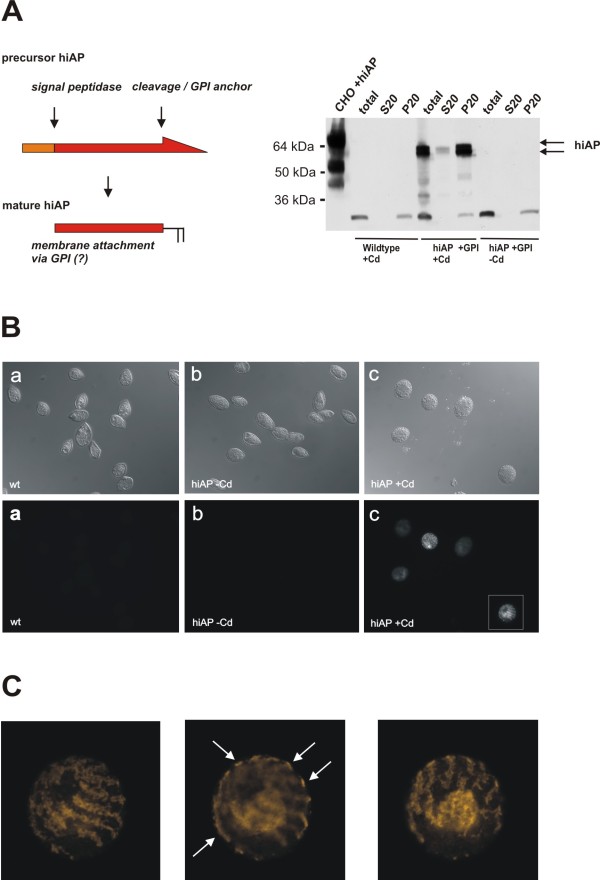
Figure 3.
Localization of recombinant hiAP in T. thermophila cells. A: Cell fractionation experiment: Cell extracts were prepared from wild type cells and hiAP expressing cells treated with cadmium (+Cd) and without cadmium (-Cd). The whole protein preparation (total) was done without detergent and subsequently centrifuged at 20,000 × g. The insoluble fraction (pellet, P20) and the soluble fraction (supernatant, S20) were analyzed by SDS-PAGE and compared to the total fraction. Almost all of the recombinant hiAP (arrows) was found in the insoluble cell fraction, suggesting that most of the recombinant protein is proper processed and therefore membrane associated. The double band probably corresponds to intracellular precursor hiAP that is membrane attached by the signal peptide or corresponds to a non-cleaved GPI anchor signal. The scheme on the left side illustrates the processing of precursor hiAP into the mature enzyme. B: Immunofluorescence analysis: In order to confirm the cell fractionation data we performed a microscopic analysis. T. thermophila cells that express hiAP and negative controls (wild type and non-induced hiAP cells) were fixed and subsequently stained by the L-19 antiserum. The upper panel shows the Nomarski images (control) of the stained cells in the lower panel. The figure clearly illustrates that only induced cells lead to a significant staining. The cell marked with a white box was analyzed in a higher magnification. C: Detailed study of image B-c: HiAP expressing cells have a distinct staining pattern. A confocal scan through the cell demonstrated that the main hiAP signals are found on the surface of the ciliate cell (arrows). Additional signals were found in the middle of the cells. This not further characterized structure probably corresponds to transport vesicles or membranous structures that carry precursor forms of the recombinant protein. Further complete detail scans of hiAP expressing cells are shown in the two Additional files 1 and 2.