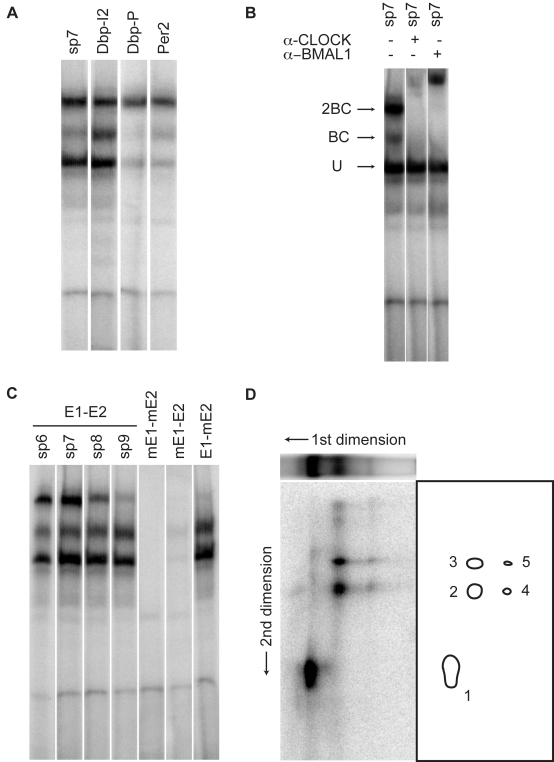
Figure 4. E1-E2 tandem sites are bound by dimers of BMAL1/CLOCK heterodimers.
(A) EMSA gel analysis with nuclear extract from mouse livers harvested at the ZT2 time point. Extracts are incubated with oligonucleotides containing the naturally occurring E1-E2 sites in the Dbp promoter (Dbp-P), Dbp intron 2 (Dbp-I2), the Per2 promoter (Per2), and an oligonucleotide with a canonical E-box (E1, CACGTG) and a non-canonical E-box (E2, AACGTG) spaced by seven nucleotides (sp7). Note: the shifts for the Dbp-I2 and sp7 probes are very similar. (B) Supershifts with anti-CLOCK and anti-BMAL1 antibodies identify two CLOCK- and BMAL1-containing complexes termed 2BC (heavier) and BC (intermediate weight), plus one unspecific complex (U, lowest weight). (C) Increased spacing and mutants. The upper 2BC band is reduced as the spacing between the E1 and E2 sites is increased from 6 bp (sp6) to 9 bp (sp9), the latter showing a pattern that resembles that obtained by mutating the E2-box but leaving E1 intact (E1-mE2). Mutating the canonical E-box (mE1-E2) strongly suppresses all complexes, while the doubly mutated probe (mE1-mE2) shows no binding. (D) Two-dimensional EMSA. The ZT2 extracts were incubated with sp7 oligonucleotides prepared with azido-dUTP nucleotides, and separated on a 1D EMSA gel (first dimension) (see Materials and Methods). The same three bands are found as in (A–C), and the weaker 2BC band compared to the regular probes (without the azido nucleotides) reflects reduced affinity following the azido substitutions in the E-box sites. In the second dimension, the five main spots indicate that the U complex contains one DNA-binding protein (spot 1), while the BC and 2BC complexes show identical DNA-binding constituents, namely CLOCK (96 kDa, spots 3 and 5) and BMAL1 (70 kDa, spots 2 and 4), as inferred by their approximate molecular mass.