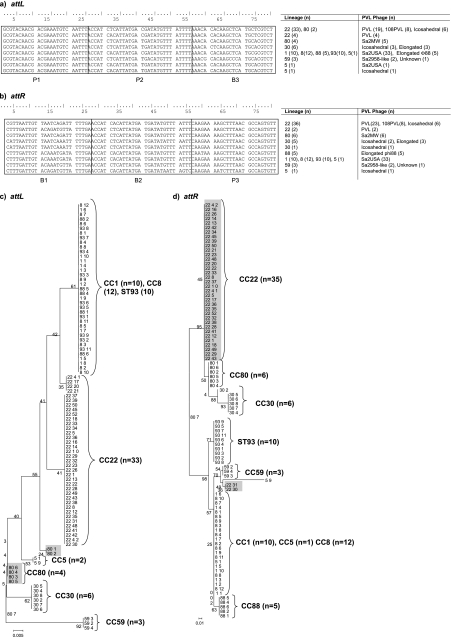
Fig. 4.
ClustalW sequence alignment of attachment motifs from 92 PVL-MRSA strains. (a) attL attachment site; (b) attR attachment site; (c and d) ML analysis tree with bootstrap values for attL (c) and attR (d). Regions of intralineage variation are shaded (CC80 and CC22). For each of the 114 PVL-MRSA isolates, the first number corresponds to the MLST clonal complex (CC) and the second to the isolate number. Branching numbers represent bootstrap values.