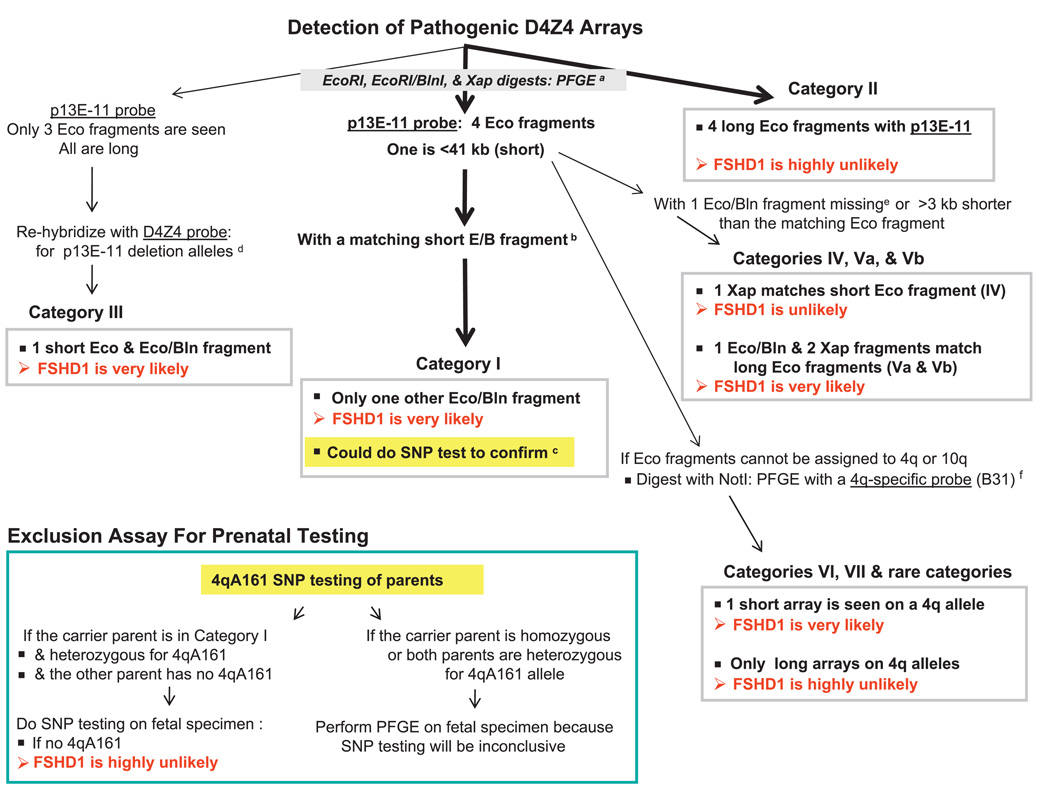
Figure 5.
A possible scheme for use of the sequencing SNP assay in clinical testing for FSHD1, which involves short 4q D4Z4 arrays and constitutes ~95% of FSHD cases.27 Analysis paths are shown by arrows in series. Thick arrows indicate most likely outcomes. aCanonical 10q D4Z4 arrays in EcoRI/BlnI digests28 and canonical 4q arrays in XapI digests are reduced to small fragments that electrophorese off the gel.9 PFGE is highly preferable to LGE, for example, PFGE can reveal somatic mosaics having five hybridising EcoRI fragments.17 18 If LGE is used, the so-called dosage test25 should be done to determine if the most proximal D4Z4 repeat unit on both 4q alleles is 4q-type (BlnIR) and on both 10q alleles is 10q-type (BlnIS), and the blot should be re-probed with D4Z4. EcoRI/HindIII digests can be used instead of EcoRI digests to get cleaner bands, but fragments are ~2 kb smaller.12 The maximum size of pathogenic D4Z4 alleles is only an approximate threshold due to incomplete penetrance, especially when EcoRI fragments are 35–41 kb; also, some patients may have very slightly larger D4Z4 arrays.33 bMatching EcoRI/BlnI fragments are ~3 kb shorter and XapI fragments are ~5 kb shorter than the corresponding EcoRI fragment.12 cThe sequencing test is the recommended assay for 4qA161, the predominant haplotype of pathogenic D4Z4 alleles. It can serve as a surrogate for 4qA/4qB testing by PFGE on HindIII digests with 4qA- and 4qB-specific oligonucleotide probes.11 20 22 23 There can be pathogenic non-4qA161 alleles with 10q-type BlnIS D4Z4 repeat units on 4q.32 However, categories I, III and VI in figure 6 do not involve such arrays; therefore, 4qA161 testing would be informative for samples giving these types of PFGE patterns, especially because ~40–50% of controls have no 4qA161 allele. dThe D4Z4 subfragment probe under stringent hybridisation conditions detects fragments with p13E-11 deletions and gives stronger signals than the p13E-11 probe from XapI digests.12 eCategories IV, Va and Vb are non-standard D4Z4 arrays with canonical 10q-type repeat units on 4q or vice versa, as evidenced by a missing EcoRI/BlnI fragment (only one EcoRI/BlnI fragment instead of two, figure 6, Vb) or an EcoRI/BlnI fragment that is less than 3 kb shorter than the corresponding EcoRI fragment (figure 6, IV and Va). fSometimes, the chromosomal origin of a short EcoRI fragment has to be determined by PFGE on NotI digests blot-hybridised to a 4q35-specific probe (B31),13 22 for example, figure 6, category VI versus category VII, and unusual cases with all four arrays beginning with 10q-type D4Z4 units. The NotI fragment length will be the sum of 187 kb and the length of the corresponding EcoRI fragment.