Fig. 2.
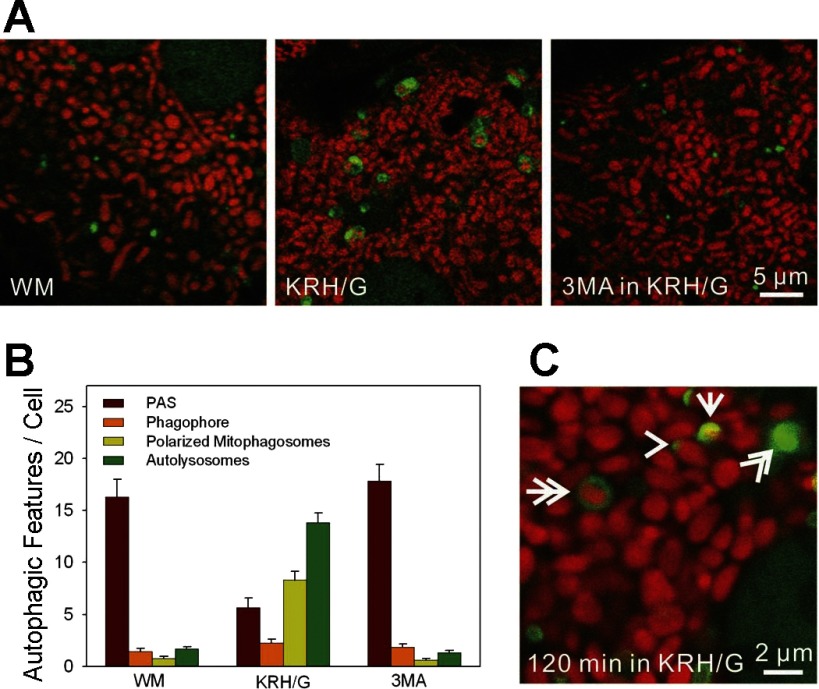
Confocal microscopy of mitophagy during nutrient deprivation plus glucagon in GFP-LC3 hepatocytes. GFP-LC3 hepatocytes were loaded with tetramethylrhodamine methylester (TMRM) to label mitochondria, as described in materials and methods. A: confocal images of TMRM-loaded GFP-LC3 hepatocytes incubated in WM (left), KRH/G (middle), or KRH/G plus 10 mM 3-methyladenine (3MA; right) for 90 min. In WM, green-fluorescing GFP-LC3 patches were present that sometimes resided adjacent to red-fluorescing mitochondria. Autophagosomes (green rings and disks) were rare but increased greatly in KRH/G and often enclosed TMRM-labeled mitochondria. 3MA blocked autophagosome formation in WM. B: the numbers of PAS (green patches), phagophores (green cup-shaped structures), polarized mitophagosomes (green rings or disks containing TMRM), and depolarized autophagosomes/autolysosomes (green rings or disks not containing TMRM) per cellular confocal image were quantified for GFP-LC3 hepatocytes incubated as described in A from three different hepatocyte isolations per treatment group. Differences in numbers of PAS, mitophagosomes, and autolysosomes in KRH/G compared with WM and 3MA were statistically significant (P < 0.05, n = 5 cells/group). C: representative structures of a PAS (arrowhead), phagophore (arrow), polarized mitophagosome (double arrow), and autolysosome (double arrowhead).