Abstract
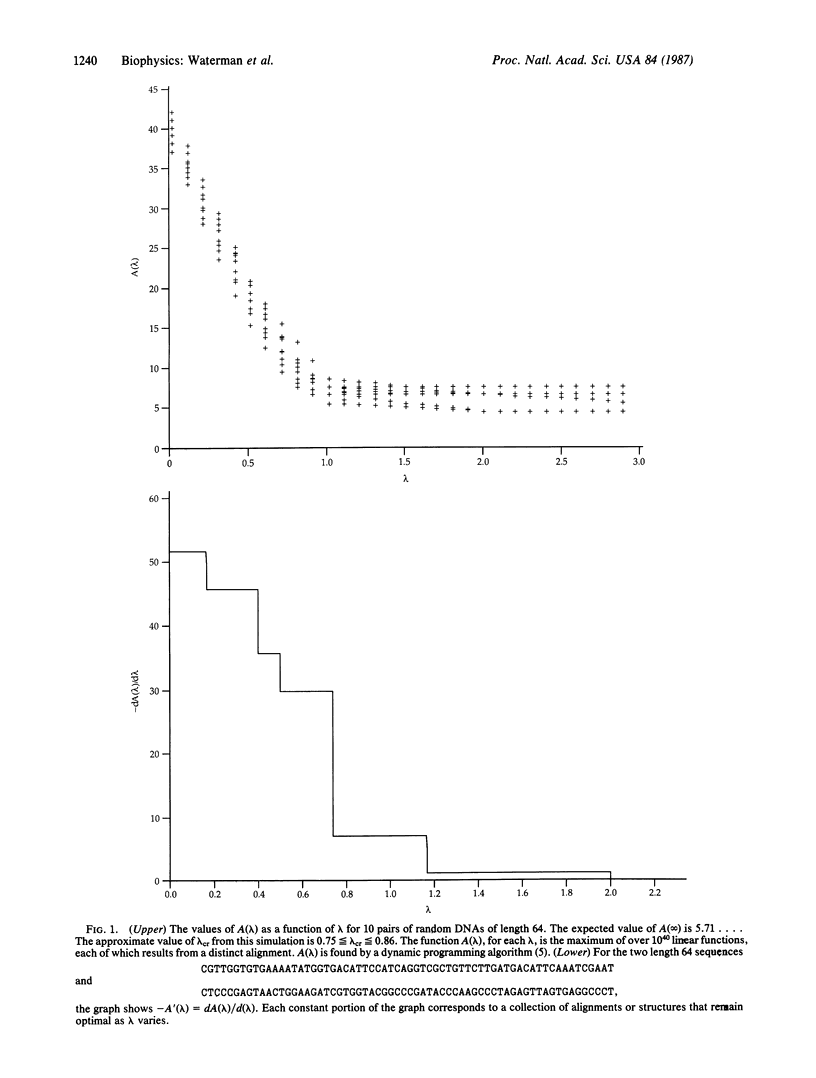
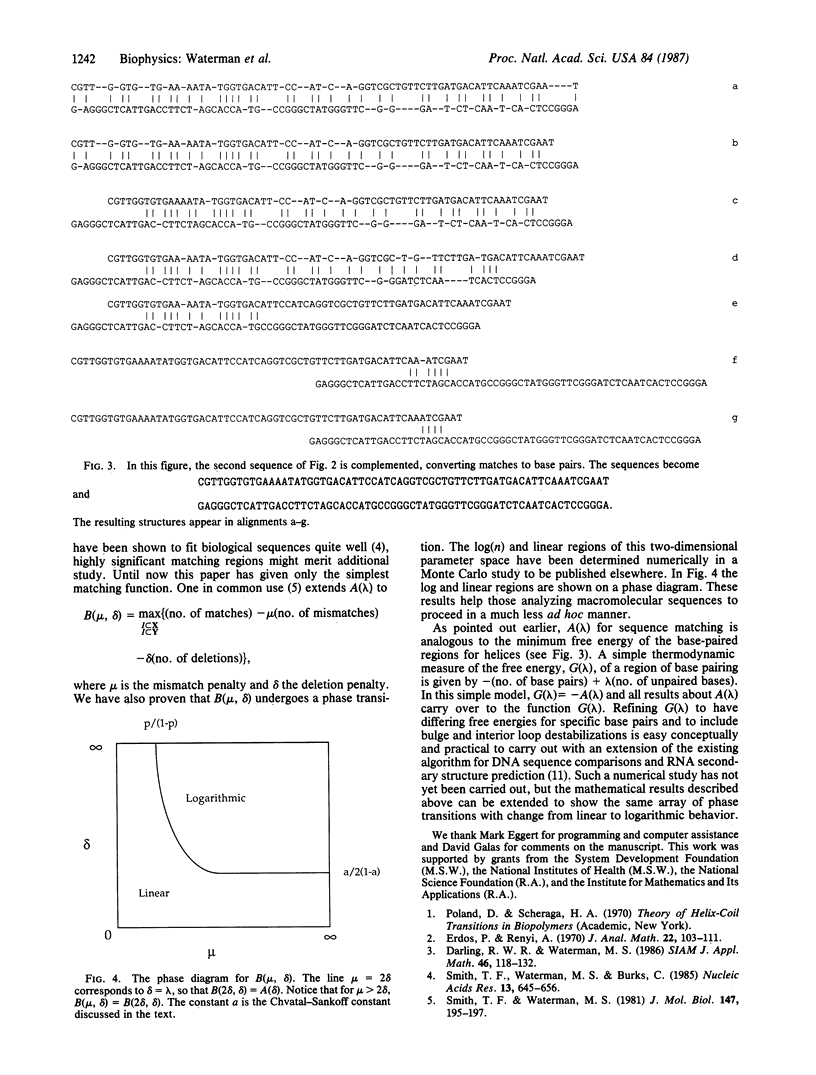
Analyses of phase transitions in biopolymers have previously been restricted to studies of average behavior along macromolecules. Extremal properties, such as longest helical region, can now be studied with a new family of probability distributions [Arratia, R., Gordon, L. & Waterman, M. S. (1986) Ann. Stat. 14, 971-993]. Not only is such extremal behavior analyzed with great precision, but new phase transitions are determined. One phase transition occurs when behavior of the free energy of the longest helical region abruptly changes from proportional to sequence length. The annealing of two single-stranded molecules and the melting of a double helix are both considered. These results, initially suggested by studies of optimal matching of random DNA sequences [Smith, T. F., Waterman, M. S. & Burks, C. (1985) Nucleic Acids Res. 13, 645-656], also have importance for significance tests in comparison of nucleic acid or protein sequences.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Karlin S., Ghandour G., Ost F., Tavare S., Korn L. J. New approaches for computer analysis of nucleic acid sequences. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5660–5664. doi: 10.1073/pnas.80.18.5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T. F., Waterman M. S., Burks C. The statistical distribution of nucleic acid similarities. Nucleic Acids Res. 1985 Jan 25;13(2):645–656. doi: 10.1093/nar/13.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T. F., Waterman M. S. Identification of common molecular subsequences. J Mol Biol. 1981 Mar 25;147(1):195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]


