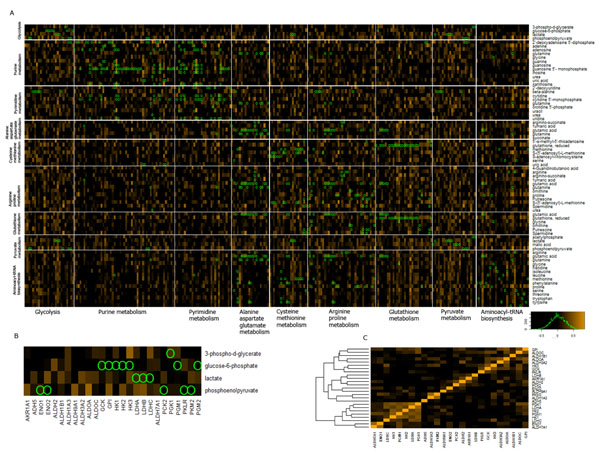
Figure 2.
Heatmap of gene-metabolite relationship organized according to KEGG pathway. Mapping of EHMN gene-metabolite association data to robust correlation matrix heat map. A. Rows are genes grouped by pathway names, columns are metabolites also grouped by corresponding pathway names. Green circles mark the position of a specific reaction that couples a metabolite and a gene from EHMN. Orange Light cells indicate positive PQC, with black to be 0 and orange to be 1. It can be seen that even though there are patterns of gene-metabolite clustering, very few high correlations can be mapped to known reactions. B. Gene-metabolite correlation in the Glycolysis pathway. C: The gene-gene correlation with in Glycolysis pathway.