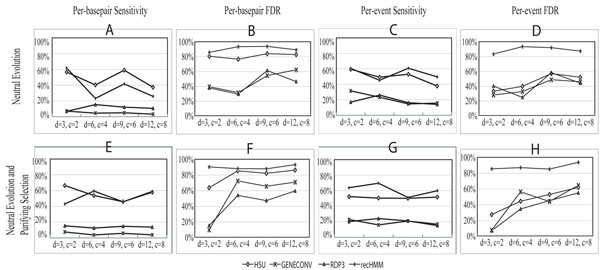
Figure 3.
Summary of the detection results. 3d duplication and 2c conversion events were simulated in each dataset, for d = {3, 6, 9, 12} and c = {2, 4, 6, 8} respectively. d duplications were simulated before the split of NWM and human, then an additional d duplications plus c conversion events were applied in each species up to the split of OWM and human. Finally, d duplications and c conversion events were applied to all species after the split of OWM and human. In A-D only neutral evolution is modeled, whereas both neutral evolution and purifying selection are included in E-H. Each point represents the mean of five replications (fewer for recHMM). (A),(E) The sensitivity of the methods for detecting converted basepairs. (B),(F) The false discovery rate (FDR) for the per-basepair detection. (C),(G) The sensitivity for detecting the existence of gene conversion events. (D),(H) The FDR for the existence detection.