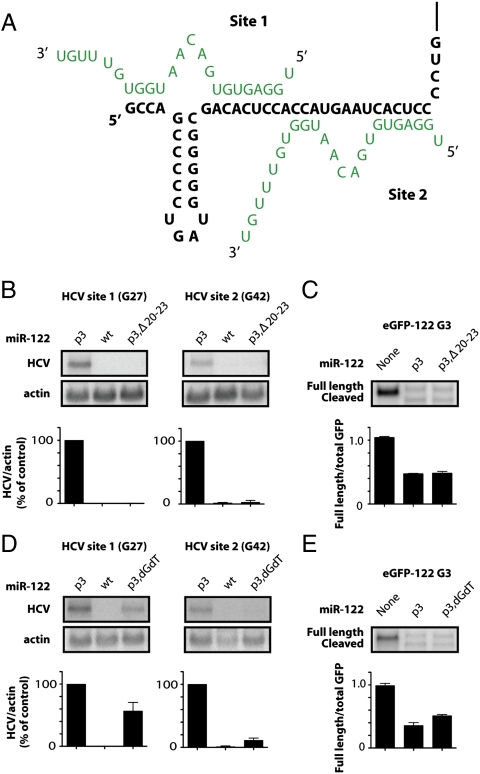
Fig. 5.
Effects of 3′ end sequences of miR-122 on HCV RNA abundance. (A) Model for miR-122 (green)-HCV (black) interactions at the 5′ end of the HCV genome. (B) Effects of 3′ end deletions in miR-122. A 19-nucleotide-long miR-122 p3 duplex (p3,Δ20–23) was targeted to site 1 or 2 in HCV RNA electroporation assays as described in Fig. 2. (C) siRNA-mediated GFP cleavage assays in HeLa cells transfected with p3,Δ20–23 as described in Fig. 3. (D) Effects of 3′ end nucleotide compositions in miR-122. The last two nucleotides of miR-122 p3 were replaced with deoxynucleotides dG and dT (p3,dGdT) and targeted to site 1 or 2 in HCV RNA electroporation assays as described in Fig. 2. (E) siRNA-mediated GFP cleavage assays in HeLa cells transfected with p3,dGdT as described in Fig. 3.