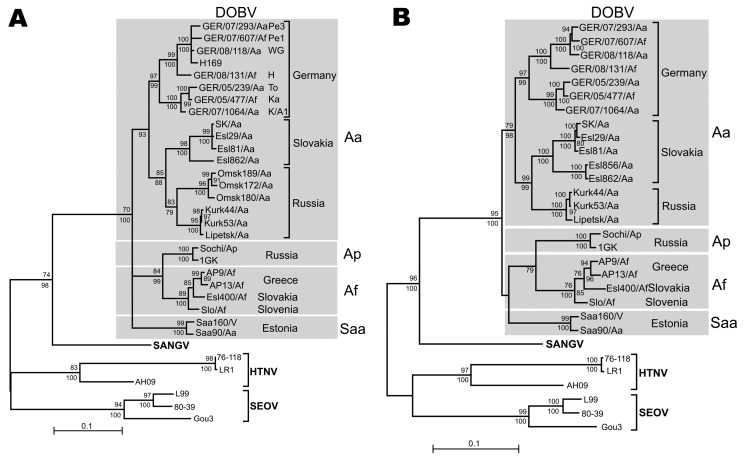
Figure 2.
Maximum-likelihood (ML) phylogenetic trees of Dobrava-Belgrade virus (DOBV) based on partial small (S) segment nucleotide sequences of 559 nt (position 377–935) (A) and complete nucleocapsid protein coding nucleotide sequences (S segment open reading frame) (B). The ML trees (Tamura-Nei evolutionary model) were calculated using TREE-PUZZLE package (www.tree-puzzle.de). Scale bars indicate an evolutionary distance of 0.1 substitutions per position in the sequence. Values above the branches represent PUZZLE support values. Values below the branches are bootstrap values of the corresponding neighbor-joining tree (Tamura-Nei evolutionary model) calculated with the PAUP* software (paup.csit.fsu.edu) from 10,000 bootstrap pseudoreplicates. Only values >70% (considered significant) are shown. Different DOBV lineages are indicated by gray boxes. SANGV, Sangassou virus; HTNV, Hantaan virus; SEOV, Seoul virus; Saa, Saaremaa virus; Aa, Apodemus agrarius; Ap, A. ponticus; Af, A. flavicollis. WG, district Lüneburg, Lower Saxony (LS); Pe1 and Pe3, district Güstrow; H, district Nordvorpommern, K/A1, district Demmin, all Mecklenburg-Western Pomerania (MWP); To and Ka, district Ostprignitz-Ruppin, Brandenburg (BB). Before tree construction, automated screening for recombination between the S segment sequences was performed using program RDP3 (15), which used 6 recombination detection programs: Bootscan, Chimeric, GENECONV, MaxChi, RDP, and SiScan with their default parameters. No putative recombinant regions could be conclusively detected by >3 programs and subsequently verified by phylogenetic trees.