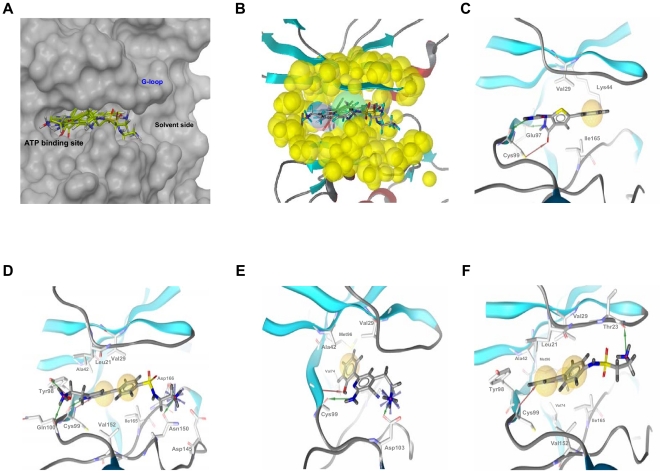
Figure 2. The ATP-binding site, the structure-based common pharmacophore and the best poses for different hIKK-2 inhibitors.
The ATP-binding site (Figure 2A), the structure-based common pharmacophore (Figure 2B) and the best poses of the various chemical hIKK-2 inhibitors that were used during the homology-model validation and the structure-based common pharmacophore construction (Figures 2C, 2D, 2E and 2F). In Figures 2C, 2D, 2E and 2F, hydrogen-bond donors, hydrogen-bond acceptors and hydrophobic interactions are shown as red arrows, green arrows and yellow spheres, respectively. Figure 2A shows the ATP-binding pocket of our hIKK-2 homology model and the best poses obtained when docking it with the hIKK-2 inhibitors 13 [27]; 12 [18]; 4a [23] and 14 [18]. Figure 2B shows the locations on the ATP-binding site of the sites that form the structure-based common pharmacophore developed in the present work and the shell of excluded volumes (in yellow) that schematically represents the locations of the surrounding residues. This pharmacophore is formed by two hydrogen-bond donors (in blue), one hydrogen-bond acceptor (in red) and one hydrophobic region (in green), with tolerances (i.e., radii) of 1.5, 1.5 and 3.0 Å, respectively. Figures 2C, 2D, 2E and 2F show the intermolecular interactions between hIKK-2 and the best poses from inhibitors 13, 12, 4a and 14, respectively. Figures 2A and 2B were drawn with Maestro v9.0.211 (Schrödinger LLC., Portland, USA; http://www.schrodinger.com), whereas Figures 2C, 2D, 2E and 2F were drawn with LigandScout v2.03.