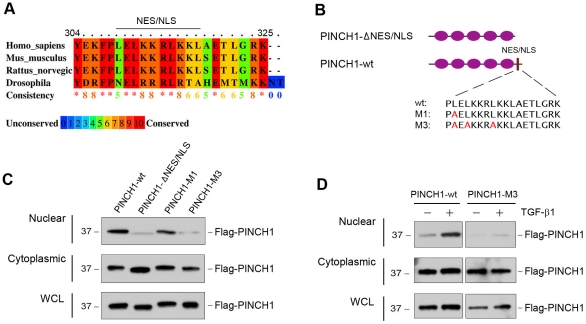
Figure 4. Subcellular localization of PINCH1 is dictated by a putative motif in its C-terminus.
A, Amino acid sequence comparison reveals a conserved, overlapped, putative NES/NLS motif in the C-terminus of PINCH1 among different species including human, mouse, rat, and Drosophila. Alignments of the deduced amino acid sequences were performed by using PRALINE program. Color bar is a strength histogram that denotes the least to most conserved amino acids (least: dark blue, light blue, green, orange, red: most). B, Schematic diagram shows the structural domains of PINCH1 and construction of various PINCH1 mutants. Purple ovals indicate the five LIM domains. The position of a putative NES/NLS motif is shown by a bar, and the sequences of wild-type and mutant NES/NLS are given. C, Deletion or mutation of the putative NES/NLS blocks nuclear translocation of PINCH1 in podocytes. Human podocytes were transfected for 48 h with Flag-tagged wild-type PINCH1 (pFlag-PINCH1-wt), truncated PINCH1 without NES/NLS (p-Flag-PINCH1-ΔNES/NLS), PINCH1 with single amino acid mutation in the NES/NLS motif (pFlag-PINCH1-M1) and PINCH1 with three amino acids mutation in the NES motif (pFlag-PINCH1-M3), respectively. Nuclear and cytoplasmic proteins were separated and immunoblotted with antibodies against Flag. D, TGF-β1 treatment fails to induce nuclear translocation of PINCH1 with mutant NES/NLS. Podocytes were transfected with wild-type PINCH1 (pFlag-PINCH1-wt) or mutant PINCH1 (pFlag-PINCH1-M3) for 48 h, respectively, and then treated TGF-β1 (2 ng/ml) for 3 h. Nuclear and cytoplasmic proteins were separated and immunoblotted with antibodies against Flag.