Abstract
Streptococcus pyogenes infections remain a health problem in several countries due to poststreptococcal sequelae. We developed a vaccine epitope (StreptInCor) composed of 55 amino acids residues of the C-terminal portion of the M protein that encompasses both T and B cell protective epitopes. The nuclear magnetic resonance (NMR) structure of the StreptInCor peptide showed that the structure was composed of two microdomains linked by an 18-residue α-helix. A chemical stability study of the StreptInCor folding/unfolding process using far-UV circular dichroism showed that the structure was chemically stable with respect to pH and the concentration of urea. The T cell epitope is located in the first microdomain and encompasses 11 out of the 18 α-helix residues, whereas the B cell epitope is in the second microdomain and showed no α-helical structure. The prediction of StreptInCor epitope binding to different HLA class II molecules was evaluated based on an analysis of the 55 residues and the theoretical possibilities for the processed peptides to fit into the P1, P4, P6, and P9 pockets in the groove of several HLA class II molecules. We observed 7 potential sites along the amino acid sequence of StreptInCor that were capable of recognizing HLA class II molecules (DRB1*, DRB3*, DRB4*, and DRB5*). StreptInCor-overlapping peptides induced cellular and humoral immune responses of individuals bearing different HLA class II molecules and could be considered as a universal vaccine epitope.
Keywords: Amino Acid, Antibodies, Antigen presentation, Lymphocyte, Peptides, NMR Structure, Structure Stability, Vaccine
Introduction
Streptococcus pyogenes causes several severe infections: acute rheumatic fever (RF)2 and rheumatic heart disease (RHD), acute glomerulonephritis, and uncomplicated pharyngitis and pyoderma. The estimated global mortality due to S. pyogenes infections around the world is ∼500,000 cases/year, and S. pyogenes is responsible for ∼233,000 RHD cases/year (1).
There are four anti-group A streptococci (GAS) vaccine candidates based on the M protein and 8 more candidates based on other streptococci antigens, such as group A CHO, C5a peptidase (SCPA), cysteine protease (Spe B), binding proteins similar to fibronectin, opacity factor, lipoproteins, Spes (super antigens), and streptococcal pili (2).
A multivalent vaccine combining the amino acid sequences of the N-terminal portion of the M protein of 26 strains of GAS, the most frequent strains in the USA, developed as a recombinant protein, is currently under phase II clinical trials (3, 4, 5).
Because the C-terminal portion of the M protein is conserved among the 200 strains identified by their emm-types, vaccines based on this region are expected to provide broad coverage. The first approach to develop a vaccine based on the C-terminal portion of the M protein was performed by Bessen and Fischetti (6), and currently his group uses the entire C-terminal portion of the M6 strain as a recombinant vaccine candidate (7). Another model uses a minimum B cell epitope (J14) from the C-terminal portion, and this vaccine candidate, currently in the experimental phase, is constructed as a chemically synthesized peptide (8, 9). Our approach is also based on the C-terminal portion of the M protein. A vaccine candidate composed of 55 amino acids was based on the reactivity of human sera and T lymphocytes against 79 overlapping peptides (10).
Vaccines are designed to present foreign antigens to the immune system to elicit a strong and long lasting protective immune response. According to the immunization summary of the World Health Organization, 3.4 million deaths were averted in the world's poorest countries through immunization programs supported by “The Global Alliance for Vaccines and Immunizations (GAVI)” between 2000 and 2008 (11). However, millions continue to die of vaccine-preventable diseases. Part of the problem is providing vaccines in a form that can be stored and remain biologically active until immunization (12). Usually, the chemical and structural integrity of the components and its immunogenic properties must be ensured through using an expensive storage, distribution and administration cold chain, critically limiting the efficacy of the immunization program in some parts of world. Vaccine stabilization strategies include screening of appropriate stabilizers and physico-chemical conditions. Currently, a more rational approach toward developing and understanding the factors affecting vaccine inactivation and stability have been adopted using spectroscopic and calorimetric techniques such as fluorescence, circular dichroism, and differential scanning calorimetry (12). This directly calls attention to the stability of vaccines with respect to temperature, pH, ionic strength, osmolarity, and the presence of excipients. Therefore, a thermodynamic and chemical stability study on the StreptInCor vaccine candidate is of great interest to determine the optimal in vitro stability conditions of the synthetic peptide. Furthermore, changes in the native conformation of the StreptInCor vaccine epitope could translate into different immunologic responses.
In this work, we present the nuclear magnetic resonance (NMR) structure of the StreptInCor vaccine polypeptide and a thermodynamic and chemical stability study of the StreptInCor folding/unfolding process using far-UV circular dichroism. These studies can be expected to provide valuable information about the structure and stability of the StreptInCor synthetic vaccine, aiming toward the small-scale processing, production, and formulation of a vaccine against S. pyogenes. Because a vaccine should provide broad coverage, we also analyzed the ability of the StreptInCor vaccine candidate to interact with different HLA class II molecules to activate the immune system and induce protective responses against GAS, regardless of the genetic background of the target population.
EXPERIMENTAL PROCEDURES
Peptide Synthesis and Purification
The vaccine candidate (StreptInCor, medical ID) was defined using a large panel of peripheral blood mononuclear cells (PBMC) from healthy individuals and RF/RHD patients (10) and consisted of 55 amino acid residues derived from the C-terminal region of the M5 protein (13) as follows: KGLRRDLDASREAKKQLEAEQQKLEEQNKISEASRKGLRRDLDASREAKKQVEKA (patents INPI 0501290/0604997–4 and ongoing registration in several countries).
Briefly, the vaccine candidate peptide (Fig. 10) and 14 overlapping peptides (numbered from 1–14, Table 1) were synthesized using solid phase technology using Fmoc (9-fluorenylmethoxycarbonyl) chemistry (14) and in situ activation of the amino acids with HOBt/HBTU (1-hydroxybenzotriazole/{N-[(1H-benzotriazol-1-yl) (dimethylamino) methylene-N-methylmethanaminium hexafluorophosphate N-oxide}. The peptide was synthesized on NovaSynTM TGR resin (Novabiochem) using DMF. The resin was contained in a fixed bed type reactor of an Advanced ChemTech robotic benchtop multiple peptide synthesis system, model APEX-396.
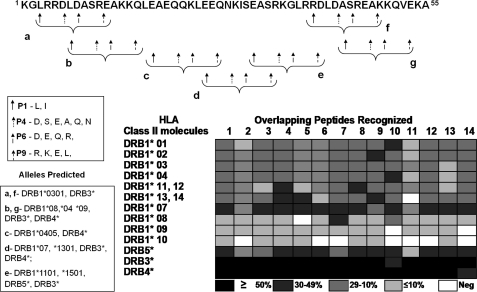
FIGURE 10.
The StreptInCor peptide is a universal vaccine epitope. A 55-residue peptide should be processed by APC to be able of bind to diverse HLA class II molecules based on the pockets (P1, P4, P6, and P9) in the HLA class II groove (18). The alternative residues of a peptide that are necessary for interactions of HLA class II alleles with the P1, P4, P6, and P9 pockets and the possibilities for StreptInCor-processed peptides to interact with several HLA class II molecules are shown in the boxes. The frequencies of recognition of the overlapping peptides according to the HLA class II alleles/molecules identified in both patients and healthy individuals showed a broad recognition spectrum of the StreptInCor vaccine epitope.
TABLE 1.
Immune recognition of overlapping M protein C-terminal peptides
| HLA |
Overlapping peptides sequences |
Individuals bearing different predicted HLA Class II allelesa with positive immune response |
|||
|---|---|---|---|---|---|
| Class II binding prediction epitopes (Fig. 10) | (20-aa residues) | Cellular response |
Humoral response |
||
| Patients 107 | Controls 90 | Patients -296 | Controls -323 | ||
| N (%) | N (%) | N (%) | N (%) | ||
| a, f, g | 1-KGLRRDLDASREAKKQLEAE | 46 (43.0) | 26 (28.9) p = 0.05 | 140 (66.5)b | 118 (81.4) p = 0.002 |
| 2-KGLRRDLDASREAKKQVEKA | 18 (16.8) | 13 (14.4) | 259 (87.5) | 265 (88.2) | |
| a, b, f, g | 3- GLRRDLDASREAKKQLEAEQ | 26 (24.3) | 12 (13.3) | 85 (40.6)b | 91 (62.8) p < 0.001 |
| 4- RRDLDASREAKKQLEAEQQK | 17 (15.9) | 14 (15.6) | 246 (83.1) | 251 (77.7) | |
| b, g | 5- LRRDLDASREAKKQLEAEQQ | 28 (20.6) | 12 (13.3) | 88 (41.9)b | 87 (60.0) p = 0.008 |
| 6- GLRRDLDASREAKKQVEKAL | 16 (15.0) | 9 (10.0) | 153 (72.9)b | 91 (62.8) p = 0.05 | |
| 7- LRRDLDASREAKKQVEKALE | 31 (29.0) | 17 (18.9) | 94 (44.8)b | 52 (36.0) | |
| c | 4- RRDLDASREAKKQLEAEQQK | 17 (15.9) | 14 (15.6) | 246 (83.1) | 251 (77.7) |
| 8- DLDASREAKKQLEAEQQKLE | 22 (20.6) | 16 (17.8) | 44 (21.0)b | 49 (33.8) p = 0.007 | |
| 9- LDASREAKKQLEAEQQKLEE | 25 (23.4) | 21 (23.3) | 50 (23.8)b | 55 (37.9) | |
| d | 10-KLEEQNKISEASRKGLRRDL | 3 (11.5)a | 4 (25.0)a | 227 (76.7) | 262 (81.1) |
| 11-KISEASRKGLRRDLDASREA | 27 (25.2) | 22 (24.4) | 232 (78.4) | 251 (77.7) | |
| e, a, f, g | 12- SEASRKGLRRDLDASREAKK | 11 (10.3) | 12 (13.3) | 232 (78.4) | 51 (77.7) |
| 13- ASRKGLRRDLDASREAKKQV | 26 (24.3) | 15 (16.7) | 232 (78.4) | 247 (76.5) | |
a 26 patients and 16 controls.
b 210 patients and 145 controls were analyzed. Overlapping peptides sequences as described (Guilherme et al., 10). T cell immune responses were considered positive when SI ≥ 2.0 (supplemental Tables S1, a and b and 2). Sera dilution to determine humoral responses was 1:100.
Preparative reverse phase chromatography was performed using a basic HPLC AKTA system (GE). The peptide molecular weight was determined using an Ettan MALDI-ToF Pro Mass Spectrometer (15).
NMR Spectroscopy
NMR spectra were recorded at 298 K on a 600-MHz Bruker spectrometer, and the data were processed using XWINNMR-update software. The NMR samples were prepared by dissolving 5 mg of the StreptInCor peptide in 300 μl of TFE-d3:H2O (30:70 (v/v)) (16). Resonance assignments were obtained from two-dimensional TOCSY (17), DQF-COSY (18), and NOESY (44) spectra, and the sequence was assigned following standard procedure (19).
NMR Structural Calculations
To obtain a structural model for the StreptInCor peptide, an ensemble of 50 structures was generated using the distance geometry-simulated annealing program DGII Φ/Ψ angles and 450 NOE-based distance restraints that were derived from NOESY spectra recorded using a 400 ms mixing time at 295 K (138 intra-residue, 222 inter-residue, and 90 overlapped restraints). The NOEs were partitioned into three categories (strong, medium, and weak) and converted into distance restraints (1.8–2.8 Å, 2.8–3.5 Å, and 3.5–5.0 Å, respectively). No coupling constants were used. All of the structures were subjected to extensive minimization, including a short molecular dynamics simulation, using the Discover program with a consistent valence force field (CVFF). The chirality of all the Cα atoms (except for glycine residues) was fixed to the L configuration, and the geometry of the peptide bonds was fixed to the trans configuration in the NMR data calculations. Hydrogen bond restraints (L24-N28) were introduced for low amide temperature coefficients. Only those <4.0 −ΔδHN/ΔT ppb/K were used in the structure calculations. Distance ranges involving these likely NH…O hydrogen bonds were set at 1.8–2.5 Å between the residue acceptor oxygens (i-4) and residue donor amide hydrogens (i).
Searching and comparison of the StreptInCor peptide model with all three-dimensional structures was carried out using the program DALI (20). The geometrical and stereochemical quality of the model was assessed using the program MOLPROBITY (21).
Circular Dichroism Structural Analysis
The secondary structure of the StreptInCor peptide was assessed using far-UV CD spectroscopy. The experiments were performed on a J-815 Jasco spectropolarimeter equipped with a nitrogen-flushed cell and a Peltier temperature control unit. CD spectra were recorded as the average of 4 or 16 repeats, using a scanning speed of 100 nm min−1, a spectral bandwidth of 1 nm, and a response time of 0.5 s. Sealed quartz cuvettes of 1-mm path length were used in all of the experiments. No loss of solvent due to evaporation at high temperature was detected. Preliminary experiments were performed under the same conditions as for the NMR experiments. The StreptInCor peptide was examined at 10 μm in 500 μl of TFE:H2O (30:70 (v/v)) at 20 °C. For thermal and chemical induced unfolding, the peptide concentrations were 78 μm and 39 μm, respectively, in PBS at pH 7.4. Changes in the ellipticity induced by the forward and reverse temperature titration were monitored at 220 nm from 4 °C to 90 °C, in increments of 2 °C at a heating rate of 20 °C h−1. The data were obtained by averaging the ellipticity values for 120 s. The chemical unfolding of the StreptInCor peptide was characterized by measuring the ellipticity change at 220 nm induced by the increase in urea concentration from 0 to 5 m at 4 °C. Because of the high amount of urea in the denatured sample, no significant data could be recorded at wavelengths below 210 nm. The ellipticity of the StreptInCor peptide at 220 nm was also monitored at 4, 18, and 36 °C as function of pH. Acetate-borate-phosphate buffer was prepared for use between pH 4 and 10, in steps of 0.5 pH units. In all of the urea denaturation and pH experiments, the samples were prepared and incubated overnight before measurement. A correction for the buffer contribution was applied to all measurements. All spectra were averaged and smoothed using a third-order Savitzky-Golay smoothing algorithm (22).
Circular Dichroism Data Analysis
For chemical denaturation, using the linear extrapolation model for two-state reversible unfolding, the free energy is linearly dependent on the denaturant concentration (23). According to this model, the free energy of unfolding in the absence of denaturant ΔGDH2O and the slope m of ΔGD as a function of the denaturant concentration can be derived from the ellipticity curves by non-linear regression. For thermal denaturation, also using the two-state reversible model, the temperature of transition Tm (i.e. the melting temperature) between the folded/unfolded states and the van 't Hoff enthalpy change ΔGDH2O can be extrapolated from the observed CD intensities by non-linear regression.
Patients and Healthy Individuals
Blood samples were collected from 296 patients with rheumatic fever/rheumatic heart disease (mean age 13.8 ± 4.8). The patients were followed for at least 5 years at the Heart Institute (InCor), HC-FMUSP, in São Paulo, Brazil. All patients were diagnosed according to the Jones criteria (24). 323 healthy subjects (mean age 37.7 ± 9.3) with no previous history of RF or any recently documented streptococcal throat infection were also included in the study. Blood-sample collection procedures were approved by the Clinical Hospital Ethics Committee, and informed consent was obtained from the parents of patients under the age of 18 years who were participating in the study.
Evaluation of the Humoral and Cellular Immune Response
The presence of antibodies and PBMC that were reactive against several overlapping peptides from which the vaccine epitope was defined, were evaluated using ELISA and proliferation assays respectively.
For ELISA, we used 96-well high binding plates (Immunoware, Pierce Biotechnology) previously coated with streptococcal M5 peptides (25 μg/ml) in 0.05 m carbonate buffer pH 9.6 for 1h at 37 °C. Sera samples were tested diluted 1:100 in 1% PBS-BSA buffer for 18 h at 4 °C. The cut-off for humoral responses was established based on the OD distribution of sera from healthy individuals and rheumatic fever patients, analyzed using the box-plot method and the Kruskal-Wallis test described and defined as the median of OD values of each peptide tested (10).
Proliferation assays were performed in 96-well plates by incubating 105 PBMC separated by density gradient centrifugation (d = 1078 g/ml) with 10 μg/ml of M protein derived-peptides for 5 days at 37 °C in a humidified 5% CO2 incubator. Negative controls were PBMC and medium in the absence of peptides and positive controls were PBMC stimulated with PHA-P (2.5 μg/ml, Sigma-Aldrich). Antigens were tested in triplicate and pulse-labeled with 0.5μCi/well of tritiated thymidine (Amersham Biosciences) in the final 18 h of culture. Cells were harvested and analyzed using a beta counter (Beta plate 1205-LKB). Proliferative response was considered as positive when the stimulation index (SI) was ≥ 2.0. SI, median of cpm of triplicate samples/median of cpm of triplicate negative control.
HLA Class II Antigens
HLA class II alleles (HLA-DR1 to DR18, HLA to DR51, DR52 and DR53) were identified for 107 patients and 90 healthy individuals using PCR-SSP methodology. DNA was isolated from proteinase K-treated peripheral blood leukocytes by salting-out extraction (25).
RESULTS
Peptide Design and Purity
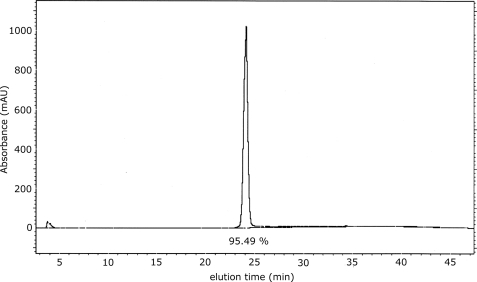
The StreptInCor vaccine candidate is a polypeptide composed of 55 amino acid residues and encompasses both T and B cell epitopes linked by 8 residues as in the natural M5 protein (10). The theoretical mass is calculated as 6361.45 Da, and the experimental mass obtained by spectrometry (MS) was 6357.5 Da. The degree of purity, as determined by HPLC, was 95.5% (Fig. 1). The resulting biopolymer has a basic theoretical pI of 9.6 (25) and an extremely low optical activity due to the absence of Trp, Tyr, and Cys residues.
FIGURE 1.
HPLC profile of the StreptInCor polypeptide. The purity of the peptide is 95.49%.
NMR Assignment
Cross-peaks between NH and CHα protons were identified by analyzing COSY spectra acquired in 30% TFE-d3/H2O. TOCSY spectra were used to correlate the side-chain spin-systems with the NH-CHα cross-peaks, following Wüthrich's (26) methodology for the assignments.
Structure Calculations
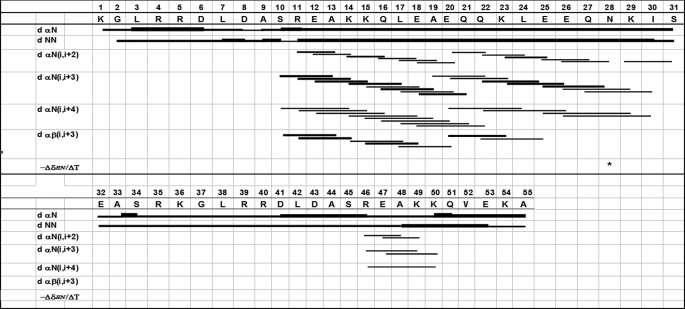
NOESY spectra for the peptide showed short and medium range sequential dNN(i,i+1), dαβ (i,i+3), dαN(i,i+3), dαN (i,i+4) NOE connectivities, suggesting the presence of a partial α-helical structure (Fig. 2). The possibility of a 310 helix was discarded due to the presence of dαN(i,i+4) NOEs and the presence of αβ (i,i+3) NOEs having medium-intensity α-helical characteristics.
FIGURE 2.
Summary of sequential medium-range NOE connectivities. The NOE intensities are represented by the line thickness. *, low coefficient, amide proton used for structure calculations.
Nine peptide structures were chosen that not present distance violations greater than 0.3 Å and ω angles greater than 1.5° out of 50 modeled structures. An average rmsd (0.56 Å) was obtained for the main-chain atoms by superimposing the structures between residues S10-I30 with a consensus structure having the least total energy. Secondary structure analysis showed the presence of an α-helix between residues E12-K29; this structural feature was confirmed by the medium-range NOEs and by the dihedral Φ and Ψ angles of each residue in the helical region adopting equal values (approximately −60° and 45°, respectively). The C- and N-terminals were flexible. A Kabsch and Sander (27) analysis of this peptide showed α-helical structure between residues E12-K29.
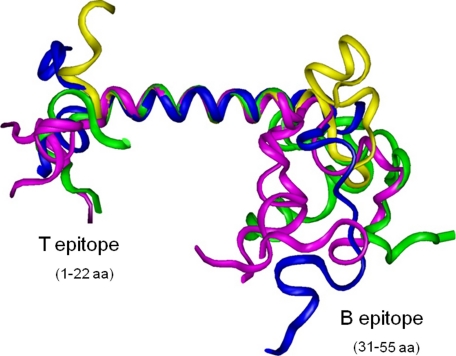
The T epitope located in the N-terminal portion of the StreptInCor peptide (residues 1–22) corresponds to the first microdomain and encompasses 11 out of the 18-residue α-helical region (Fig. 3). The second domain corresponds to the B epitope (residues 31–55) and showed no α-helical structure. Residues Lys-23 to Lys-29, which are in an α-helical structure, correspond to the 7 out of 8 amino acids residues that are located between the T and B epitopes as previously defined (10).
FIGURE 3.
Superimposition of the backbone atoms in the 9 conformers representing the NMR ensemble structure of the StreptInCor peptide. The structure shows two microdomains linked by an α-helix. The first microdomain plus the first 11 residues of the α-helix region correspond to the T epitope (25-mer) on the left side. The second domain on the right side corresponds to the B epitope (22-mer).
Structure Stability of StreptInCor: CD Spectroscopy
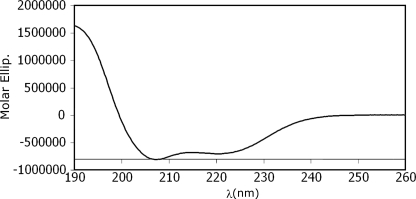
While evaluating the secondary structure, the CD spectra in TFE/H2O showed a double minima at 208 and 222 nm and a maximum at 195 nm, clearly indicating an α-helical configuration (28) (Fig. 4). This conclusion was corroborated further by the 1H-NMR studies.
FIGURE 4.
Far-UV CD spectra of the StreptInCor polypeptide in 30% TFE. The form of the curve shows the typical behavior of an α-helix.
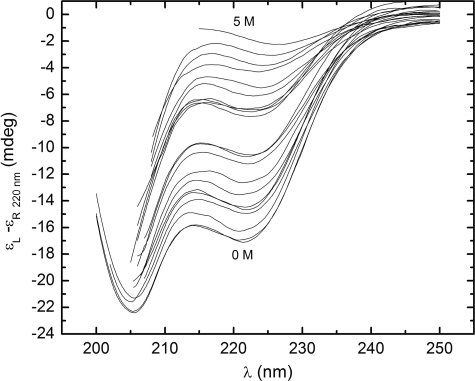
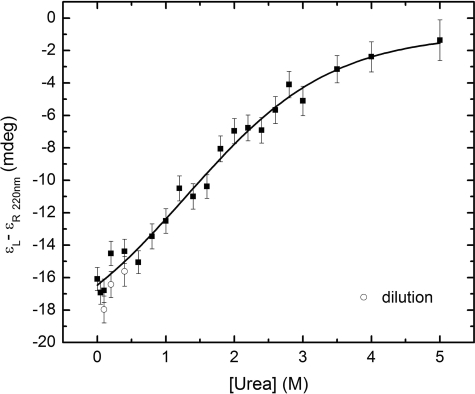
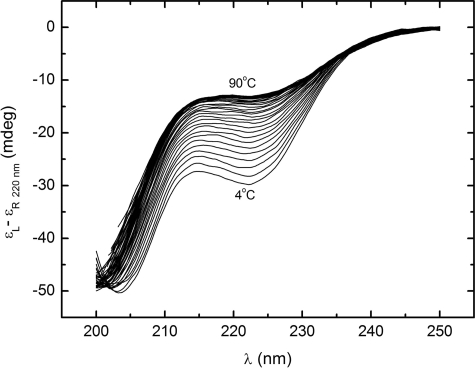
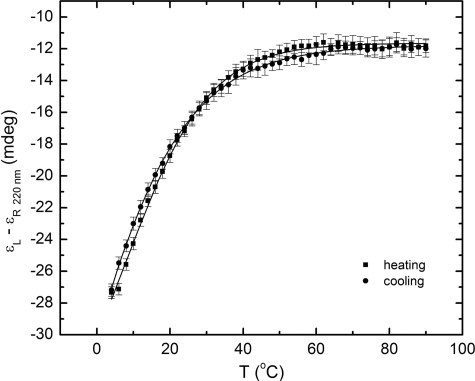
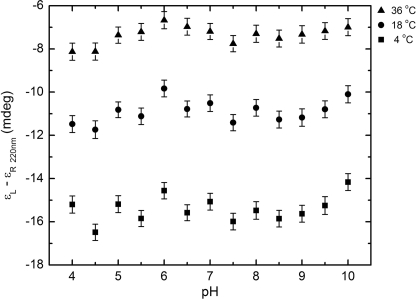
The urea-induced denaturation CD spectra of the StreptInCor peptide are shown in Fig. 5. The series of curves evinces a loss of helical content with increasing urea concentration. The ellipticity minimum at 220 nm decreases while the minimum at 205 nm is displaced to the noise region of the spectra below 200 nm. This occurs because of increasing disorder of the residues. The far-UV CD spectrum of the StreptInCor peptide in 5 m urea shows an absence of secondary structure elements. Fig. 6 presents the overall ellipticity of StreptInCor peptide monitored at 220 nm. The ellipticity decreases almost linearly between 0 and 2.6 m urea and then changes slowly until 5 m is reached, where no further variation is observed. The urea denaturation profile of the StreptInCor peptide follows a single transition between 0 and 2.4 m, and the concentration that leads to half-denaturation ([U]1/2) is 1.5 m. Samples of Streptincor peptide at 312 μm in 1, 2, and 3 m of urea were diluted 10 times, yielding the same molar ellipticities as 31 μm samples prepared in 100, 200, and 300 mm of urea. Thus, the chemical unfolding of StreptInCor peptide can be considered a reversible process. Assuming a two-state model, the free energy of unfolding of StreptInCor calculated from linear extrapolation is 1.2 ± 0.6 kcal mol−1 and the linear coefficient m of the free energy as a function of the urea concentration is 0.5 ± 0.1 kcal mol−1. The smoothed CD spectra of the StreptInCor peptide taken at different temperatures ranging from 4 °C to 90 °C (using 2 °C increments) are shown in Fig. 7. The helicity decreased as the temperature increased, as manifested by the loss of the minimum ellipticity around 220 nm and by the shift of the minimum ellipticity at 205 nm to 195 nm. The peptide ellipticity at 220 nm decreased by 20 mdeg from 4 °C to 90 °C. Almost no secondary structure is present at the higher temperature. Fig. 8 shows the overall ellipticity of the StreptInCor peptide monitored at 220 nm for the forward and reverse temperature between 4 °C and 90 °C. The ellipticity decreased rapidly from 4 °C to 30 °C and changed slowly until 58 °C, at which temperature ∼99.8% of the peptide (based on a plot of the fraction denatured) was unfolded. No variation in ellipticity was observed upon heating further. The overall ellipticity decayed as the temperature increased, and the presence of an isodichroic point at 201 nm indicated a state transition. The increasing and decreasing temperature titration curves superimposed well. These results revealed that the thermal unfolding of the StreptInCor peptide proceeds as a two-state, reversible process. This model is also corroborated by the thermodynamic parameters derived from both the forward and reverse temperature titrations. The van 't Hoff enthalpy change ΔGDH2O determined for StreptInCor is 16.4 ± 0.6 kcal mol−1 K−1 for thermal unfolding and 14.2 ± 0.5 kcal mol−1 K−1 for the reverse process, yielding an average value of 15.3 ± 0.5 kcal mol−1 K−1. The melting temperature Tm of the StreptInCor peptide unfolding process was 4 ± 1 °C and 3 ± 2 °C (for unfolding and refolding, respectively), yielding an average melting temperature of 4 ± 2 °C. No significant variation of StreptInCor peptide ellipticity was observed between pH 4 and 10, even in temperatures well above the melting temperature of the peptide. Fig. 9 shows the ellipticity of StreptInCor measured at 220 nm at different pH values at 4, 16, and 36 °C. The correlation coefficients of the three data sets were found to be close to 1.0, thus the observed variation in ellipticity was negligible.
FIGURE 5.
Urea-induced titrations in the far-UV CD spectra of StreptInCor. The urea concentration increases from 0 m (lower curve) to 5 m (upper curve).
FIGURE 6.
Overall ellipticity at 220 nm of StreptInCor at 4 °C as a function of the concentration of urea. The non-linear regression fit is indicated by the continuous line.
FIGURE 7.
Temperature-induced titrations in the far-UV CD spectra of StreptInCor. The temperature increases from 4 °C (lower curve) to 90 °C (upper curve).
FIGURE 8.
The overall ellipticity of StreptInCor at 220 nm as a function of the temperature. Symbols used: temperature increasing from 4 °C to 90 °C (■); temperature decreasing from 90 °C to 4 °C (○). The non-linear regression fit is indicated by the continuous line.
FIGURE 9.
The overall ellipticity of StreptInCor at 220 nm as a function of pH. No variation in ellipticity was observed upon pH titrations.
Humoral and Cellular Immune Response to the StreptInCor Vaccine Epitope
The StreptInCor vaccine epitope (55-mer) encompasses two repeated blocks of amino acids (KGLRRDLDASREAKKQ) along its sequence (residues 1–16 and 36–50, Fig. 10A) due to structural characteristic of the C-terminal region of the M-protein (29). We analyzed both the cellular and humoral immune response against 13 different StreptInCor-derived overlapping peptides that have been numerically identified in Table 1. We observed varied frequency in the positive T cell immune response (from 10.3 to 43.0%) in RF/RHD patients and the controls (107 and 90 samples, respectively) (Table 1 and supplemental Tables S1, a and b, and S2). Seven peptides (1, 2, 3, 5, 6, 7, and 13) had 14 residues in common (LRRDLDASREAKKQ). However, RF/RHD patients preferentially recognized the peptide 1 (p = 0.05). All individuals, as expected, presented high T cell immune response against PHA, a polyclonal mitogen (supplemental Tables S1, a and b and S2). The presence of specific antibodies against the 13 overlapping peptides was detected in the sera of a large panel of RF/RHD patients and controls (296 and 323 samples, respectively) (Table 1). Peptides 1, 3, 5, and 8 were preferentially recognized by healthy individuals (p < 0.01) and peptide 6 by patients (p = 0.05).
Considering the broad recognition of the StreptInCor epitope by both T cells and antibodies, we theoretically predicted the binding of the processed peptides (13–15 mers) to different HLA class II molecules and their fit in the P1, P4, P6, and P9 pockets in the groove of several HLA class II molecules (30). We found several peptides (a–f) that were possible candidates to fit in the groove of several class II molecules (Fig. 10). Considering the presence of repeated blocks of amino acids (KGLRRDLDASREAKKQ) along the StreptInCor peptide sequence (residues 1–16 and 36–50), as mentioned above, we identified identical potential for processed peptides to bind to HLA class II molecules (a and f, and b and g; Fig. 10).
We identified HLA class II alleles of patients and controls who were tested for cellular immune responses against the StreptInCor overlapping peptides (Table 1), and the frequencies of the allele distribution are shown in Table 2. Both DRB1*07 and DRB3 alleles were more frequent in patients (p = 0.002, O.R. = 2.99; and p = 0.01 O.R. = 0.39, respectively), confirming our previous results (31). However, the DRB1*09 allele was absent in the patient group (p = 0.002, O.R. = 0.05). The other HLA class II alleles showed no statistical differences.
TABLE 2.
Frequency of HLA class II antigens
| HLA class II antigens | Patients (107) | Controls (90) | p value |
|---|---|---|---|
| N (%) | N (%) | ||
| DRB1*01 | 19 (17.8) | 12 (13.3) | |
| DRB1*02 | 33 (30.8) | 19 (21.1) | |
| DRB1*03 | 23 (21.5) | 19 (21.1) | |
| DRB1*04 | 23 (21.5) | 21 (23.3) | |
| DRB1*11 and *12 | 20 (18.7) | 25 (27.8) | |
| DRB1*13 and *14 | 34 (31.8) | 35 (38.9) | |
| DRB1*07 | 38 (35.5) | 14 (15.6) | p = 0.002, OR = 2.99 |
| DRB1*08 | 7 (6.5) | 8 (8.9) | |
| DRB1*09 | 0 | 8 (8.9) | p = 0.05, OR = 0.05 |
| DRB1*10 | 6 (5.6) | 4 (4.4) | |
| DRB5* (DR51) | 33 (30.8) | 19 (21.1) | |
| DRB3* (DR52) | 77 (72.0) | 78 (86.7) | p = 0.01, OR = 0.39 |
| DRB4* (DR53) | 61 (57.0) | 43 (47.8) |
Considering all processing for StreptInCor of being processed and that diverse generated peptides could be presented to the T cell receptor (TCR) resulting in B cell activation, we analyzed the recognition frequencies of the 13 overlapping peptides (Table 1) based on the HLA class II alleles/molecules of both patients and healthy individuals (Fig. 10). Our results using proliferation assays showed that most of the individuals respond to several StreptInCor overlapping peptides (Fig. 10). Although the DRB1*10 allele presented low frequencies in both patients and controls (Table 2), individuals bearing this molecule are able to recognize 7 out of the 14 overlapping peptides (Fig. 10).
DISCUSSION
Vaccine design is complex and researchers should attempt to discover vaccines that are immunogenic for the global population without deleterious reactions. The StreptInCor candidate vaccine described herein has been designed based on sequences from the M5 protein (13) located in the C2 and C3 repeat domains that are conserved in all group A streptococci (32).
The three-dimensional structural features of StreptInCor are favorable for MHC class II and TCR-peptide interactions and strongly indicate the possibility of the StreptInCor vaccine epitope being recognized by professional antigen-presenting cells (APCs) of individuals bearing any HLA class II molecules with consequent activation of T cells, which undergo further maturation from effector T cells to memory cells. It is interesting to note that the T-epitope of StreptInCor originating from the C2-repeat region of the M5 protein has an α-helical structure located in the first domain of the vaccine epitope, followed by seven amino acid residues that are also in α-helical structure originated from a non-repeated region and is characteristic of the M5-type. StreptInCor also displays a B cell epitope that originates from the C3-repeat region of the M protein. This epitope is located in the second microdomain of StreptInCor, and has been shown by NMR structure to have no alpha helical conformation, indicating direct antibody-antigen interactions that lead to the possibility of antigen recognition and the induction of memory B cells.
To acquire an effective immune response against a pathogen such us S. pyogenes, both humoral and cellular immune responses are required. The results presented here fulfill this requirement and show broad immune responses mediated by both specific antibodies and T cells, and probably memory cells against StreptInCor overlapping peptides that favor a protective effect for this vaccine candidate. The T helper immune response is very important in a vaccine, as recently discussed by Kauffmann (33), and will induce both central memory T cells (in the lymph nodes) and effector memory T cells (in tissues). The central memory T cells replicate efficiently and activate B cells to produce specific antibodies that are responsible for the first antibody burst against the pathogen (this work, S. pyogenes) (33).
The major concern when developing a vaccine against S. pyogenes is avoid inducing deleterious reactions that could lead to autoimmune reactions such as rheumatic fever and their sequels (principally, those driven to the heart valves and acute post-streptococcal glomerulonephritis, in susceptible individuals). The M protein is considered the major antigen of the group A streptococci, and its structure and amino acid sequence were defined several years ago (29, 34). This led to the characterization of both polymorphic (N-terminal) and conserved (C-terminal) regions (29).
Pathogen epitopes, mainly from the N-terminal region that present structural or sequential similarity to self-epitopes, might activate autoreactive T lymphocytes that escape immune tolerance by the molecular mimicry mechanism. These autoreactive T cells can also activate B cells that will produce pathogen- and self-antigen-specific antibodies. We (35, 36) and others (37, 38) have described epitopes derived from the N-terminal portion of the streptococcal M protein capable of activating specific T cells that are cross-reactive with self-peptides. All the results have shown that defined regions of the streptococcal M5 protein (i.e. the N- terminal region) play a role in the development of inflammatory heart disease, leading to myocarditis and valvulitis in humans. However, knowledge acquired about the mechanism that leads to pathogenic autoimmunity and the identification of streptococcal M protein pathogenic epitopes that are recognized by B and/or T cells have allowed the search for that protective epitopes that have been presented here. The StreptInCor vaccine candidate has been tested in several animal models and no pathogenic reactions were observed.
Bessen and Fischetti (6) have defined M protein C-terminal peptides capable of inducing IgA antibodies and reported the prevention of bacterial colonization in animal models. Following these experiments, a live mucosal vaccine was engineered to express C-repeat M protein epitopes. This vaccine, using the commensal organism Streptococcus gordonii, was developed and tested in mice (7). This commensal vector was evaluated as a vaccine delivery vehicle, and preliminary results showed that the vector was safe and well tolerated when tested by oral and nasal routes in 150 healthy volunteers (39). Another anti-streptococcal vaccine model that was based on a combination of C- and N-terminal peptides from Australian and New Zealand aboriginal disease-related S. pyogenes strains is being assayed (40). These authors identified a C-terminal epitope called P145 (20-mer) that elicited a protective antibody response in a murine model (41). However, they found that human T cells recognized both the P145 peptide and some amino acid residues of human myosin (42). To avoid any deleterious reaction, they constructed several chimeric peptides using a protein derived from yeast and several P145-overlapping peptides that were named as J1 to J14 (41). In vivo challenge experiments confirmed the protective efficacy of immunization with the J14 peptide in different formulations (42, 43, 9). In addition, the vaccine potential of J8 (one of the chimeric peptides) was evaluated in murine models and passed to clinical phase I studies currently underway (44). A fundamental step in the development of vaccines is to discover epitopes able to induce immune protective responses independently of the genetic background of such populations.
The role of class I and II HLA molecules is to present antigens to the T cell receptor of CD8+ and/or CD4+ T cells, triggering the activation of the adaptive immune responses via T and B antigen-specific cells. The prediction that the StreptInCor vaccine candidate can be processed into different peptides and is capable of binding to any HLA class II molecules suggests broad capacity of the immunization coverage by the vaccine epitope. The fact that StreptInCor-overlapping peptides were recognized by both T cells and antibodies lends support to this statement and suggests that the vaccine epitope is able to induce a broad protective immune response.
The StreptInCor peptide adopts a molecular structure showing no significant structural similarity with any known three-dimensional structures in the Protein Data Bank. Its NMR chemical shifts in aqueous solution clearly deviate from those observed for unstructured/disordered peptides. The StreptInCor macromolecular structure is composed of two microdomains linked by an 18-residue α-helix from Arg-11 to Ser-34. The N-terminal microdomain encloses the first 22 residues of the T epitope and the next 11 residues of the central α-helix. Most of the remaining residues adopt a loop conformation, and a few residues, Arg-5-Asp-8, adopt a bend conformation. Residues Lys-23 to Lys-29 adopt an α-helical conformation. However, these do not account for any of the epitopes. The second microdomain, the B epitope, is composed of two bends between residues Arg-35-Asp-41 and Ala-44-Arg-46, a helical turn between Glu-47-Lys-50, and the remaining residues, which adopt a loop conformation. The StreptInCor model presented very good stereochemical parameters. Most of the amino acid residues have their main-chain Φ/Ψ angles lying in favored regions of the Ramachandran plot. Only 2 out of 54 (3.7%) residues were found in disallowed regions. Hydrophobic interactions between the adjacent residues Leu-7 and Arg-4 may explain the unfavorable Φ/Ψ angles of D6. The other residue with Φ/Ψ angles disallowed position is Gly-37, which lies between two tightly packing residues, Arg-39 and Lys-36. Only 3 residues out of 46 (6.5%) were found to have a poor rotamer conformation, and all of these are lysine residues (Lys-14, Lys-36, and Lys-54) that are hidden from solvent. Finally, the model has 3 Cβ deviations (>0.25 Å), and there are no bond length or bond angle violations.
The StreptInCor polypeptide fulfills all of the requirements for thermodynamic analysis with a two-state reversible model. Both reversibility and monomolecularity were found to be true. The melting curves of StreptInCor measured using different concentrations are superimposable, indicating that the peptide is monomolecular. In addition, the StreptInCor chemical and thermal folding/unfolding behavior proceeds as a two-state reversible process without any intermediate states. StreptInCor is chemically stable, as expected for covalently bound peptide chain. Regarding its structural stability, almost no variation was detected in its helicity over a broad pH range. When in the presence of a strong denaturant, the peptide reversibly unfolds almost spontaneously at moderate urea concentrations as a consequence of its low free energy. The secondary structure of StreptInCor is susceptible to variations in temperature and presents a relatively low melting temperature. Specifically, under in vitro conditions, about half of the peptide molecules are unfolded at 4 °C. The difference between the forward and reverse melting temperatures might be related to differences in the initial and final peptide conformations or to thermal stress effects. Nevertheless, in regard to peptide structural stability, such temperature sensitivity is unimportant as long as it can be reverted to its most stable conformational state by lowering the temperature upon storage. The van't Hoff enthalpy change describes the amount of heat released or absorbed during the course of a reaction, as determined indirectly by measuring the temperature dependence of the equilibrium constant. The van't Hoff enthalpy change determined for the StreptInCor peptide was very close to that observed for the helix-coil transition of a 50-residue peptide in water (45). Taking into account only the 18 residues that adopting an α-helical conformation, the enthalpy change of StreptInCor is 0.85 kcal mol−1 K per residue, in excellent agreement with the predicted value (0.86 kcal mol−1 K) for several alanine-rich helices (46). Taken together, these results show that StreptInCor satisfies the necessary conditions regarding chemical and structural stability, and therefore it can be considered an appropriate reagent for the development of an anti-streptococcal vaccine for preventing rheumatic fever and other sequelae of S. pyogenes infections.
In conclusion, the structural, chemical, and biological properties presented here for StreptInCor, an anti-S. pyogenes vaccine candidate polypeptide, indicate that it can be considered as a natural and universal vaccine against severe streptococcus-induced diseases.
Supplementary Material
This work was supported by “Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP)” and “Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq),” Brazil and Colombia.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2.
- RF
- rheumatic fever
- RHD
- rheumatic heart disease
- NMR
- nuclear magnetic resonance
- TFE
- trifluoroethanol
- TCR
- T cell receptor
- APC
- antigen-presenting cell
- GAS
- group A streptococci
- PBMC
- peripheral mononuclear cells
- SI
- stimulation index.
REFERENCES
- 1. Carapetis J. R., Steer A. C., Mulholland E. K., Weber M. (2005) Lancet. Infect. Dis. 5, 685–694 [DOI] [PubMed] [Google Scholar]
- 2. Steer A. C., Batzloff M. R., Mulholland K., Carapetis J. R. (2009) Curr. Opin. Infect. Dis. 22, 544–552 [DOI] [PubMed] [Google Scholar]
- 3. Hu M. C., Walls M. A., Stroop S. D., Reddish M. A., Beall B., Dale J. B. (2002) Infect. Immun. 70, 2171–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McNeil S. A., Halperin S. A., Langley J. M., Smith B., Warren A., Sharrat G. P., Baxendale D. M., Reddish M. A., Hu M. C., Stroop S. D., Linden J., Fries L. F., Vink P. E., Dale J. B. (2005) Clin. Infect. Dis. 41, 1114–1122 [DOI] [PubMed] [Google Scholar]
- 5. Dale J. B., Penfound T., Chiang E. Y., Long V., Shulman S. T., Beall B. (2005) Clin. Diagn. Lab. Immunol. 12, 833–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bessen D., Fischetti V. A. (1988) Infect. Immun. 56, 2666–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Medaglini D., Pozzi G., King T. P., Fischetti V. A. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 6868–6872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brandt E. R., Sriprakash K. S., Hobb R. I., Hayman W. A., Zeng W., Batzloff M. R., Jackson D. C., Good M. F. (2000) Nat. Med. 6, 455–459 [DOI] [PubMed] [Google Scholar]
- 9. Batzloff M. R., Yan H., Davies M. R., Hartas J., Lowell G. H., White G., Burt D. S., Leanderson T., Good M. F. (2005) J. Infect. Dis. 192, 1450–1455 [DOI] [PubMed] [Google Scholar]
- 10. Guilherme L., Faé K. C., Higa F., Chaves L., Oshiro S. E., Freschi de Barros S., Puschel C., Juliano M. A., Tanaka A. C., Spina G., Kalil J. (2006) Clin. Dev. Immunol. 13, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. WHO - World Health Organization (2008) The World Health Report 2008: Primary Health Care Now More Than Ever, World Health Organization, Lisboa, Porto [Google Scholar]
- 12. Brandau D. T., Jones L. S., Wiethoff C. M., Rexroad J., Middaugh C. R. (2003) J. Pharm. Sci. 92, 218–231 [DOI] [PubMed] [Google Scholar]
- 13. Robinson J. H., Atherton M. C., Goodacre J. A., Pinkney M., Weightman H., Kehoe M. A. (1991) Infect. Immun. 59, 4324–4331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M. R., Appel R. D., Bairoch A. (2005) in Protein Identification and Analysis Tools on the ExPASy Server (Walker J. M. ed) Humana Press, Totowa, NJ [Google Scholar]
- 15. Guilherme L., Postol E., Freschi de Barros S., Higa F., Alencar R., Lastre M., Zayas C., Puschel C. R., Silva W. R., Sa-Rocha L. C., Sa-Rocha V. M., Pérez O., Kali J. (2009) Methods 316–321 [DOI] [PubMed] [Google Scholar]
- 16. Roccatano D., Colombo G., Fioroni M., Mark A. E. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 12179–12184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bax A., Davis D. G. (1985) J. Magn. Reson. 65, 355–360 [Google Scholar]
- 18. Rance M., Sørensen O. W., Bodenhausen G., Wagner G., Ernst R. R., Wüthrich K. (1983) Biochem. Biophys. Res. Commun. 117, 479–485 [DOI] [PubMed] [Google Scholar]
- 19. Jeener J., Meier B. H., Backman P., Ernst R. R. (1979) J. Chem. Phys. 71, 4546–4553 [Google Scholar]
- 20. Holm L., Sander C. (1993) J. Mol. Biol. 233, 123–138 [DOI] [PubMed] [Google Scholar]
- 21. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic. Acids Res. 35, Web Server issue, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Savitzky A., Golay M. J. E. (1964) Anal. Chem. 3, 1627–1639 [Google Scholar]
- 23. Pace C. N. (1986) Methods Enzymol. 131, 266–280 [DOI] [PubMed] [Google Scholar]
- 24. Dajani A. S., Taubert K. A., Takahashi M., Bierman F. Z., Freed M. D., Ferrieri P., Gerber M., Shulman S. T., Karchmer A. W., Wilson W. (1994) Circulation 89, 916–922 [DOI] [PubMed] [Google Scholar]
- 25. Olerup O., Zetterquist H. (1992) Tissue Antigens 39, 225–235 [DOI] [PubMed] [Google Scholar]
- 26. Wüthrich K. (1986) NMR of Proteins and Nucleic Acids, Wiley, New York, NY [Google Scholar]
- 27. Kabsch W., Sander C. (1983) Biopolymers. 22, 2577–2637 [DOI] [PubMed] [Google Scholar]
- 28. Segrest J. P., Jones M. K., De Loof H., Brouillette C. G., Venkatachalapathi Y. V., Anantharamaiah G. M. (1992) J. Lipid Res. 33, 141–166 [PubMed] [Google Scholar]
- 29. Fischetti V. A., Jones K. F., Hollingshead S. K., Scott J. R. (1988) Rev. Infect. Dis. 10, Suppl. 2, S356–S359 [DOI] [PubMed] [Google Scholar]
- 30. Stern L. J., Brown J. H., Jardetzky T. S., Gorga J. C., Urban R. G., Strominger J. L., Wiley D. C. (1994) Nature. 368, 215–221 [DOI] [PubMed] [Google Scholar]
- 31. Guilherme L., Weideback W., Kiss M. H., Snitcowsky R., Kalil J. (1991) Circulation. 83, 1995–1998 [DOI] [PubMed] [Google Scholar]
- 32. Smeesters P. R., Mardulyn P., Vergison A., Leplae R., Van Melderen L. (2008) Vaccine. 26, 5835–5842 [DOI] [PubMed] [Google Scholar]
- 33. Kauffman S. H. (2007) Nat. Rev. Microbiol. 5, 491–50417558425 [Google Scholar]
- 34. Manjula B. N., Trus B. L., Fischetti V. A. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 1064–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Guilherme L., Cunha-Neto E., Coelho V., Snitcowsky R., Pomerantzeff P. M. A., Assis R. V., Pedra F., Neumann J., Goldberg A., Patarroyo M. E., Pillegi F., Kalil K. (1995) Circulation 92, 415–420 [DOI] [PubMed] [Google Scholar]
- 36. Guilherme L., Oshiro S. E., Faé K. C., Goldberg A. C., Tanaka A. C., Kiss M. H., Silva C., Guzman F., Patarroyo M. E., Southwood S., Sette A., Kalil J. (2001) Infect. Immun. 69, 5345–5351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cunningham M. W., Antone S. M., Smart M., Liu R., Kosanke S. (1997) Infect. Immun. 65, 3913–3923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cunningham M. W. (2000) Clin. Microbiol. Rev. 13, 470–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kotloff K. L., Wasserman S. S., Jones K. F., Livio S., Hruby D. E., Franke C. A., Fischetti V. A. (2005) Infect. Immun. 73, 2360–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pandey M., Sekuloski S., Batzloff M. R. (2009) Immunol. and Cell Biol. 87, 391–399 [DOI] [PubMed] [Google Scholar]
- 41. Pruksakorn S., Currie B., Brandt E., Phornphutkul C., Hunsakunachai S., Manmontri A., Robinson J. H., Kehoe M. A., Galbraith A., Good M. F. (1994) Int. Immunol. 6, 1235–1244 [DOI] [PubMed] [Google Scholar]
- 42. Shaila M. S., Nayak R., Prakash S. S., Georgousakis M., Brandt E., McMillan D. J., Batzloff M. R., Pruksakorn S., Good M. F., Sriprakash K. S. (2007) Vaccine 25, 3567–3573 [DOI] [PubMed] [Google Scholar]
- 43. Vohra H., Dey N., Gupta S., Sharma A. K., Kumar R., McMillan D., Good M. F. (2005) Res. Microbiol. 156, 575–582 [DOI] [PubMed] [Google Scholar]
- 44. Olive C., Hsien K., Horváth A., Clair T., Yarwood P., Toth I., Good M. F. (2005) Vaccine 23, 2298–2303 [DOI] [PubMed] [Google Scholar]
- 45. Scholtz J. M., Marqusee S., Baldwin R. L., York E. J., Stewart J. M., Santoro M., Bolen D. W. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 2854–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ooi T., Oobatake M. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 2859–2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.