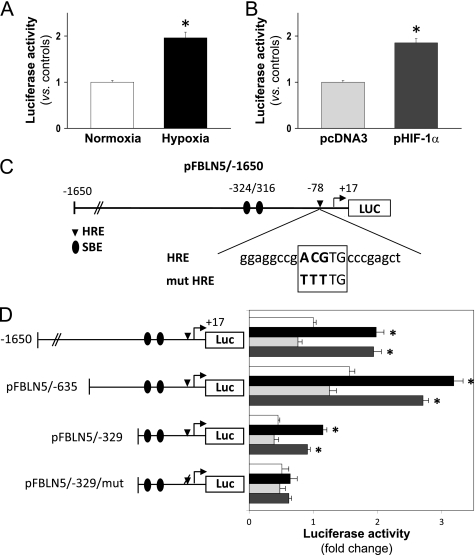
FIGURE 5.
Hypoxia up-regulates FBLN5 transcriptional activity through an HRE. A, BAEC transfected with the pFBLN5–1650 luciferase (Luc) construct were exposed to normoxia (white bars) or hypoxia (black bars) for 18 h. B, alternatively, cells maintained under normoxic conditions were co-transfected with an HIF-1α expression vector (pHIF-1α; dark gray bars) or the corresponding empty vector (pcDNA3; light gray bars). Luciferase and β-galactosidase activities were determined as described under “Experimental Procedures.” Results are expressed as mean ± S.E. (n = 9). C, scheme corresponding to the FBLN5 promoter region analyzed in transient transfection studies. The location of two Smad binding sites (SBE) and the sequence of the putative HRE motif are shown. Changes introduced by mutagenesis are indicated in boldface. D, a promoter serial deletion study was performed using the wild-type pFBLN5-1650, pFBLN5-635, and pFBLN5-329 luciferase constructs or the HRE-mutated vector pFBLN5-329/mut. Position of the wild-type HRE motif (indicated as a diamond) or its mutated form (indicated as a deleted diamond) are shown. Cells were cultured under normoxia (white bars) or hypoxia (black bars), co-transfected with a HIF-1α expression vector (dark gray bars) or the corresponding empty vector (pcDNA3; light gray bars). Luciferase activity is expressed as fold-change using pFBLN5-1650 (in normoxia) as a reference value. Results are expressed as mean ± S.E. (n = 9; *, p < 0.05 versus normoxic or pcDNA3-transfected cells).