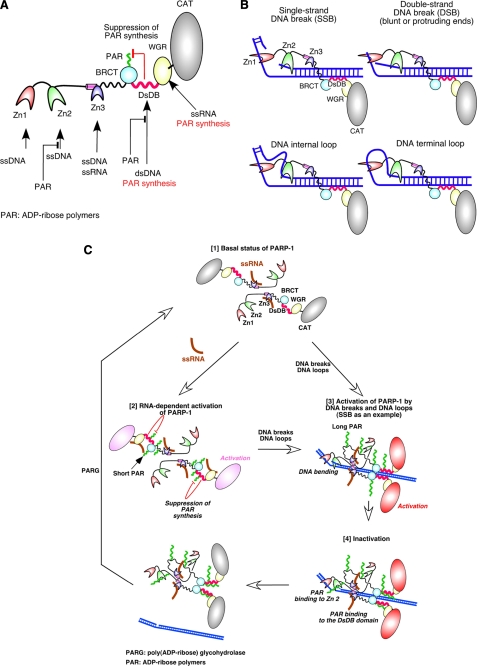
FIGURE 7.
A model for the binding of the DBD domain to DNA breaks and DNA loops and the regulatory mechanisms of PARP-1 activity. A, a summary of our results is shown. Zn1 and Zn2 bind to ssDNA. Zn3 has the ability to bind to ssRNA in addition to ssDNA. The DsDB domain is involved in recognition of dsDNA. When the domain does not bind to dsDNA, it suppresses ADP-ribose polymer synthesis. By binding of this domain to dsDNA, the suppression is removed. The WGR domain binds to ssRNA, resulting in activation of the CAT domain. Upon the formation of ADP-ribose polymers, ssDNA and dsDNA are displaced from Zn2 and the DsDB domain, respectively, by the polymers. B, a model for the DBD domain binding to DNA breaks and DNA loops is illustrated. Zn1 binds to one DNA strand, and Zn2 holds the complementary strand of SSBs. Because Zn1 and Zn2 are connected by a peptide of only 20 residues, binding of both fingers to DNA strands could bend DNA. In a similar manner, Zn1 and Zn2 can bind to DSBs, DNA internal loops, and DNA terminal loops. C [1], in the basal status, poly(ADP-ribosyl)ation activity of PARP-1 is suppressed by other PARP-1 domains. The DsDB domain is one of such domains, which has role in inhibition of long ADP-ribose polymer formation. Two PARP-1 molecules are illustrated. [2], binding of ssRNA to the WGR domain activates the CAT domain. However, only short polymers are produced due to the suppression of ADP-ribose polymer synthesis by the DsDB domain. [3], when Zn1 and Zn2 bind to a SSB, PARP-1 forms a functional homodimer. PARP-1 is aligned with DNA strand through binding of the DsDB domain to dsDNA, leading to the activation of the CAT domain. Binding of the DsDB domain to dsDNA releases the suppression of ADP-ribose polymer formation, allowing the CAT domain to produce long ADP-ribose polymers. PARP-1 could bind to DSBs and to DNA loops in a similar manner to as to SSBs. [4], ADP-ribose polymers then displace ssDNA and dsDNA from Zn2 and the DsDB domain, respectively, resulting in dissociation of PARP-1 from DNA breaks. ADP-ribose polymers are then degraded by poly(ADP-ribose) glycohydrolase (PARG).