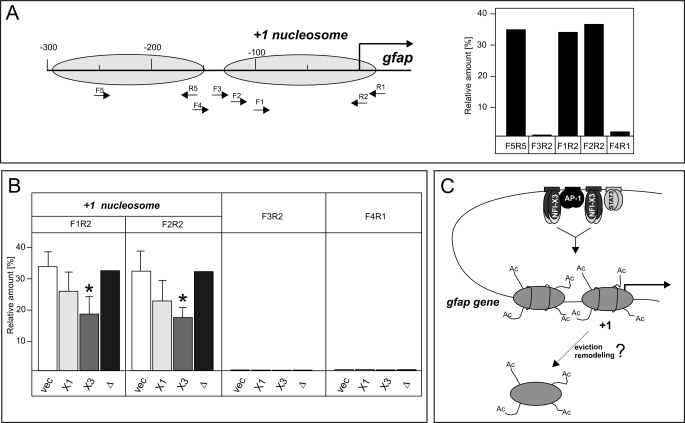
FIGURE 9.
NFI-X3 induces alterations in the nucleosome architecture at the gfap promoter. Nuclei were isolated from U373-Vec, U373-NFI-X1, U373-NFI-X3, and U373-NFI-X3Δ cells and digested with MNase as described under “Experimental Procedures.” A, the positions of mapped −1 and +1 nucleosomes are indicated, and locations of qPCR primers are shown. qPCRs were performed on both MNase-treated and total genomic DNA of U373-Vec cells, and the differences in Ct values are shown relative to the −3kb qPCR product (Ct values for −3kb were identical for genomic and MNase-treated DNA). B, qPCRs were performed on MNase-treated DNA of U373-Vec (vec), U373-NFI-X1 (X1), U373-NFI-X3 (X3), and U373-NFI-X3Δ (Δ) cells as indicated, and the differences in Ct values are shown relative to the −3kb qPCR product. Experiments were performed four times except for U373-NFI-X3Δ cells (one experiment). C, model of transcriptional activation of the gfap gene expression by NFI-X3. DNA is demethylated at the gfap enhancer, which allows for binding of the AP-1, NFI-X1/NFI-X3, and STAT3 in U373-NFI-X1/NFI-X3 cells. At the gfap promoter, histones are similarly acetylated in U373-NFI-X1 and U373-NFI-X3 cells. However, the unique TA domain of NFI-X3 induces stronger alteration of the nucleosome architecture (either remodeling or eviction) that allows for both the increased PolII occupancy and transcription of the gfap gene. Error bars, S.D.