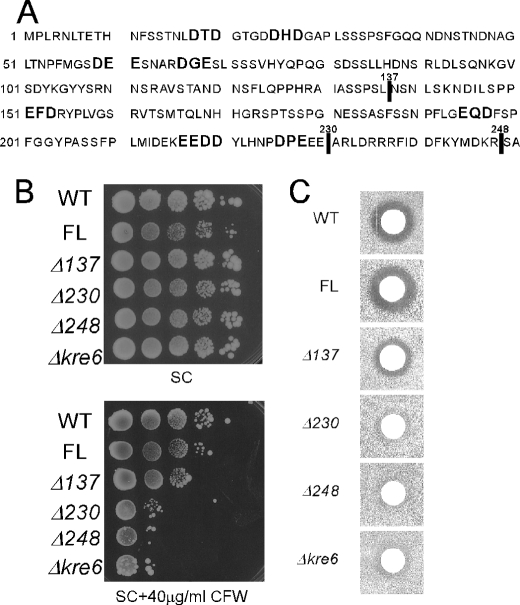
FIGURE 8.
Deletions including possible ER exit motifs in the cytoplasmic domain and the biological activity of Kre6. A, the amino acid sequence of the cytoplasmic domain of Kre6 is shown with indication of di-acidic sequences. Vertical lines indicate the ends of deletions. B, Calcofluor White sensitivity of the wild-type (WT) and Δkre6 mutant with chromosomal integration of various KRE6–3HA derivatives (full-length (FL) or three truncated versions) at the ura3-52 locus are shown. Cells were grown in SC medium until A600 nm = 1, diluted 2-fold, and then serially diluted 4 times by 10-fold. Ten μl of these samples were spotted on SC plates with or without 40 μg/ml Calcofluor White (CFW) and incubated at 30 °C for 3 d. C, K1 killer toxin sensitivity of the strains were used in B. The cells were grown in YPD at 30 °C until A600 nm = 1.0 and plated on a low-pH YPD plate. Five μl of a fresh overnight culture of the K1 killer producer was spotted, and the plates were incubated at 25 °C for 2 days to show the growth inhibitory zone.