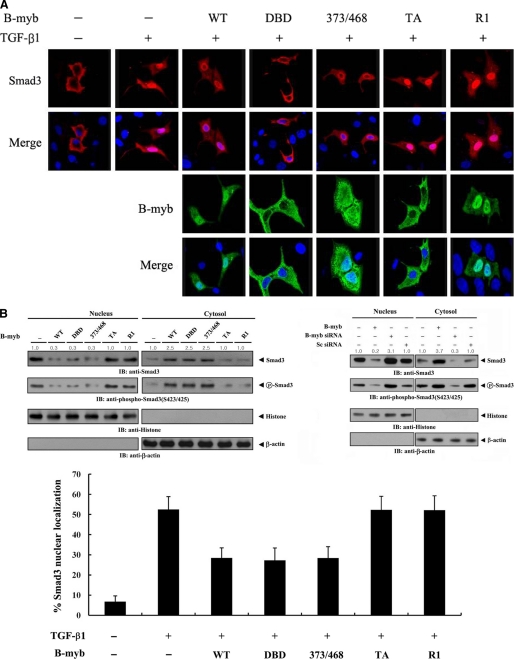
FIGURE 5.
The effect of B-MYB on the subcellular localization of SMAD3. A, modulation of SMAD3 localization by B-MYB. Hep3B cells were transiently transfected with a plasmid vector expressing FLAG-SMAD3, together with GFP-tagged wild-type B-MYB or a deletion mutant (DBD, 373/468, TA, and R1), and incubated in the presence or absence of 100 pm TGF-β1. Cells were immunostained with anti-FLAG antibody, followed by Alexa Fluor-594 anti-mouse secondary antibody (red, for SMAD3), and analyzed by confocal microscopy. B-MYB subcellular localization was determined by immunostaining with anti-GFP and Alexa Fluor-488 anti-rabbit secondary antibodies (green, for B-MYB). Nuclei (blue) were stained with DAPI. B, cytoplasmic and nuclear fractions from the extracts of Hep3B cells transfected with the indicated plasmid vectors or siRNA duplexes were separated from each other, incubated with TGF-β1, and used for immunoblot analysis with the indicated antibodies. Nucleus and Cytosol indicate the nuclear and cytoplasmic fractions of cell extracts, respectively. The relative level of SMAD3 expression was quantified by densitometric analysis. The fold-increase relative to untransfected samples in parental Hep3B cells was calculated. The percentage of SMAD3 nuclear localization was calculated as the number of SMAD3-positive cells with nuclear localization divided by the total number of SMAD3-positive cells (lower panel). About 280 cells were analyzed per sample using a confocal microscopy.