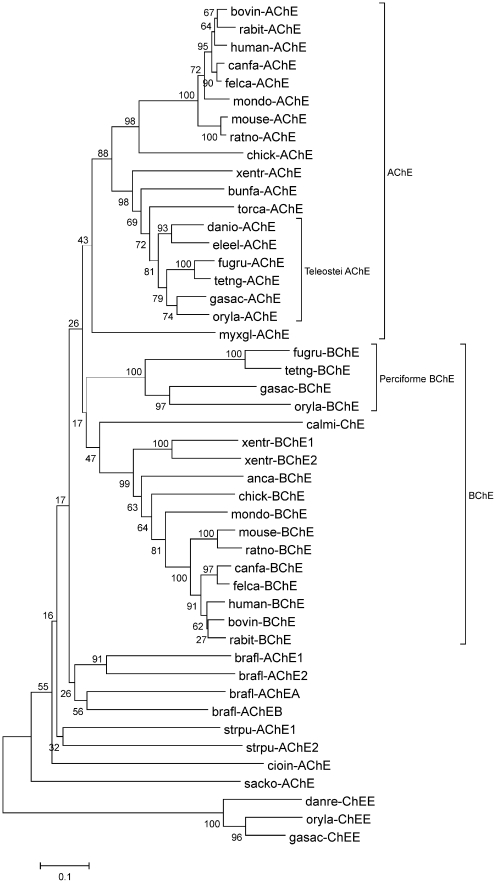
Figure 2. Evolutionary relationships of 47 taxa.
The evolutionary history was inferred using the Neighbor-Joining method. The optimal tree with the sum of branch length = 8.65 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method [102] and are in the units of the number of amino acid substitutions per site. All positions containing gaps and missing data were eliminated from the dataset (Complete deletion option). There were a total of 257 positions in the final dataset. Phylogenetic analyses were conducted in MEGA4 [103]. Abbreviations and references found in ESTHER [104]. Mammals: bovin (Bos taurus) cattle, rabit (Oryctolagus cuniculus) rabbit, human (Homo sapiens), canfa (Canis familiaris) dog, felca (Felis catus) cat, mondo (Monodelphis domestica) possum, mouse (Mus musculus), ratno (Rattus norvegicus). Birds: chick (Gallus gallus) chicken. Amphibians: xentr (Xenopus tropicalis) toad. Reptiles: bunfa (Bungarus fasciatus) snake, anca (Anolis carolinensis) anole. Bony fish (teleosts): danio, danre (Danio rerio) zebrafish, eleel (Electrophorus electricus) electric eel, fugru (Fugu rubripes) puffer fish, tetng (Tetraodon nigroviridus) puffer fish, gasac (Gasterosteus aculeatus) stickleback, oryla (Oryzias latipes) medaka, calmi (Callorhyncus milii) elephant fish. Cartilaginous fish: torca (Torpedo californica) electric ray. Jawless fish: myxgl (Myxine glutinosa) hagfish. Urochordates: cioin (Ciona intestinalis) sea squirt, oikdi (Oikopleura dioica) tunicate appendicularium. Cephalochordates: brafl (Branchiostoma floridae) amphioxus. Hemichordates: sacko (Saccoglossus kowalevskii) acorn worm. Echinoderms: strpu (Strongylocentrus purpuratus) sea urchin.