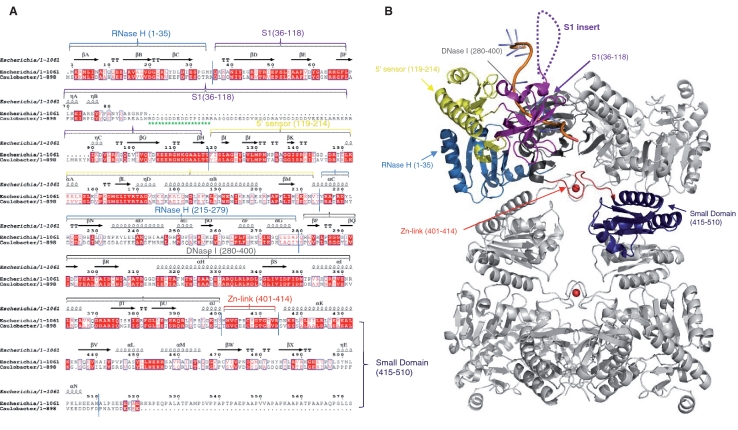
Figure 1.
RNase E of α-proteobacteria have a distinguishing S1 domain insert not found in RNase E of other bacterial classes. (A) Structure based sequence alignment of the E. coli and C. crescentus RNase E catalytic domains. The secondary structural elements of E. coli RNase E are shown on the lines above the sequence alignment using the PDB file 2BX2. The arrows indicate β-sheet, the coils indicate α-helices, TT indicates β turns and η indicates 310 helices. Red letters indicate homology and blue boxes show similarity. The red highlights indicate identity across the sequences. The green stars represent the antigenic peptide used in this study. Structural sub-domains of RNase E are coloured (RNase H: light blue; S1: purple; 5′ sensor: yellow; DNase I: dark grey; zinc link: red; small domain: dark blue). Alignments were prepared using CLUSTALW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html) and ESPript (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi). (B) E. coli RNase E NTD tetramer in complex with 13-mer RNA (pale orange). Structural sub-domains are highlighted for one protomer, coloured as in Figure 1A. The position of the S1 insert absent in E. coli RNase E catalytic domain is represented by the dashed loops.