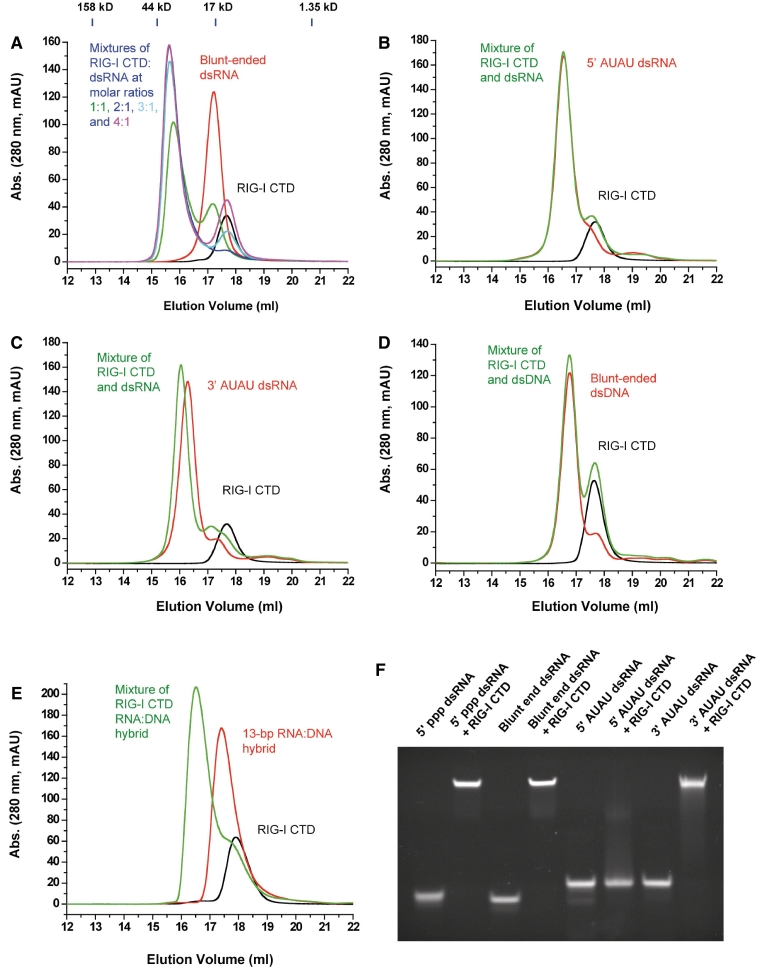
Figure 1.
RIG-I CTD binds blunt-ended dsRNA without 5′ ppp. (A) Binding studies of RIG-I CTD with the 14-bp blunt-ended dsRNA by gel filtration chromatography. RIG-I CTD, the 14-bp blunt-ended dsRNA and mixtures of RIG-I CTD:dsRNA at molar ratios 1:1 to 4:1 were injected over a superdex200 column. Elution profile of RIG-I CTD is in black and the dsRNA in red. Elution profiles of mixtures of RIG-I CTD and the 14-bp dsRNA at molar ratios 1:1, 2:1, 3:1 and 4:1 are in green, blue, cyan and purple, respectively. The molecular masses of four protein standards and their elution positions are shown above the chromatograms. (B) Binding studies of RIG-I CTD with a 14-bp dsRNA containing 5′ AUAU overhangs. A 2:1 mixture of RIG-I CTD and the dsRNA was analyzed by gel filtration chromatography. (C) Binding studies of RIG-I CTD with a 14-bp dsRNA containing 3′AUAU overhangs. The molar ratio between RIG-I CTD and the dsRNA is 2:1 in the sample. (D) Binding studies of RIG-I CTD with a 14-bp blunt-ended dsDNA with the same sequence as the 14-bp blunt-ended dsRNA. The molar ratio between RIG-I CTD and the dsDNA is 2:1. (E) Binding studies of RIG-I CTD with a 13-bp RNA:DNA hybrid. The molar ratio between RIG-I CTD and the RNA:DNA hybrid is 2:1. (F) Binding studies of RIG-I CTD with the 14-bp 5′ ppp dsRNA, 14-bp blunt-ended dsRNA and 14-bp dsRNA with either 5′ or 3′ overhangs by EMSA.