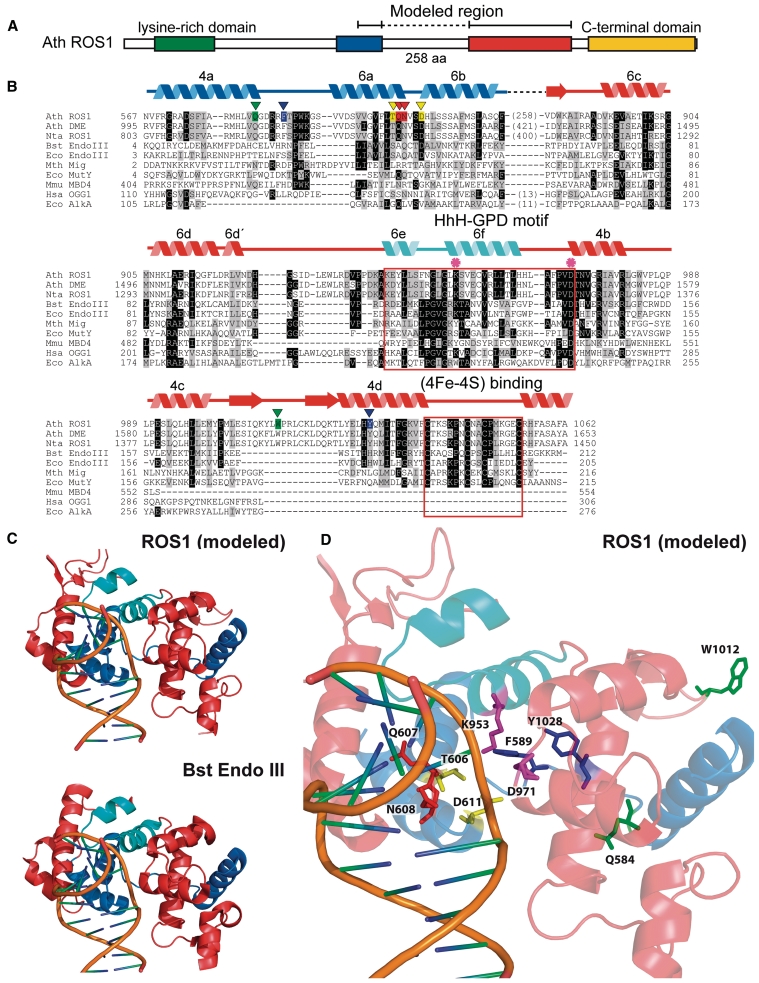
Figure 1.
An unusual sequence insertion is present in the DNA glycosylase domain of members of the DML family. (A) Schematic diagram showing ROS1 regions conserved among DML proteins. (B) Multiple sequence alignment of DML proteins and several HhH-GPD superfamily members. Listed above the primary sequence are indicated secondary structure assignments from the ROS1 model prediction shown in (C), colored according to regions shown in (A). The helix–hairpin–helix of the HhH-GPD motif is shown in cyan. ROS1 amino acids mutated in this study are indicated by inverted triangles and highlighted in green (Q584 and W1012), blue (F589 and Y1028), yellow (T606 and D611) or red (Q607 and N608). The lysine residue that is diagnostic of bifunctional glycosylase/lyase activity, and the conserved aspartic acid residue in the active site are indicated by asterisks. The HhH-GPD and the [4Fe–4S] cluster loop (FCL) motifs are boxed. Names of organisms are abbreviated as follows: Ath, Arabidopsis thaliana; Nta, Nicotiana tabacum; Bst, Bacillus stearothermophilus; Eco, Escherichia coli; Mth, Methanobacterium thermoautotrophicum; Mmu, Mus musculus; Hsa, Homo sapiens. Genbank accession numbers are: Ath ROS1: AAP37178; Ath DME: ABC61677; Nta ROS1: BAF52855; Bst EndoIII: 1P59; Eco EndoIII: P20625; Mth Mig: NP_039762; Eco MutY: NP_417436; Mmu MBD4: 1NGN; Hsa OGG1: O15527; Eco AlkA: P04395. (C) Ribbon diagrams of the structural model for the DNA glycosylase domain of ROS1 and the crystallographic Bst EndoIII structure used as template. Structural elements are colored as in (A). The duplex DNA is shown in orange. Nucleic acid coordinates extracted from the Bst EndoIII-DNA trapped complex were used to superimpose a DNA structure with a flipped-out AP site analog onto the ROS1 model. (D) Close-up view of the ROS1 model. Mutated residues are shown as sticks and colored according to (B). The conserved lysine and aspartic acid residues are shown in magenta.