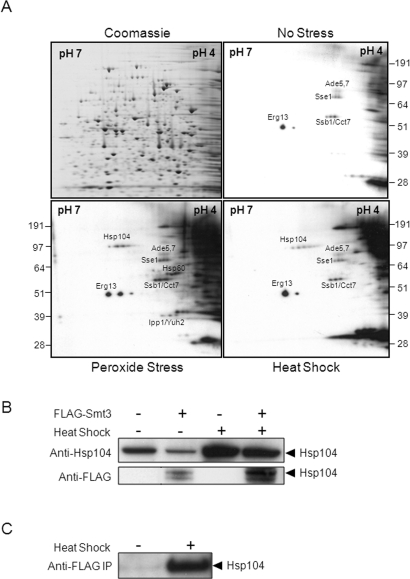
FIGURE 2:
Identification of sumoylated proteins in C. albicans using a proteomic screen. (A) Cells expressing FLAG-Smt3 (MLC01) were grown for 5 h and then exposed to stress for 1 h. Protein extracts were prepared, run on replicate 2-D gels, and either stained with Coomassie blue or subjected to Western blotting with an α-FLAG antibody: Western blots of no-stress control, peroxide-treated cells (50 mM H2O2), and heat-shocked cells (30°C to 42°C). Autoradiographs were aligned with the Coomassie-stained gels, spots chosen for analysis, and the corresponding proteins identified by tryptic digestion and LC-MS/MS. The identities of some sumoylation targets are shown. (B) THE1 cells (FLAG-Smt3 −) and MLC01 cells (FLAG-Smt3 +) were heat shocked for 1 h, analyzed by Western blotting with an anti-Hsp104 antibody, and compared with untreated cells. Membranes were then reprobed with an anti-FLAG antibody. The band corresponding to the molecular mass of Hsp104 is highlighted. (C) Untreated and heat-shocked MLC01 cells were immunoprecipitated with an anti-FLAG antibody, and lysates were analyzed by Western blotting with the anti-Hsp104 antibody. The highlighted band corresponds to the molecular mass of Hsp104.