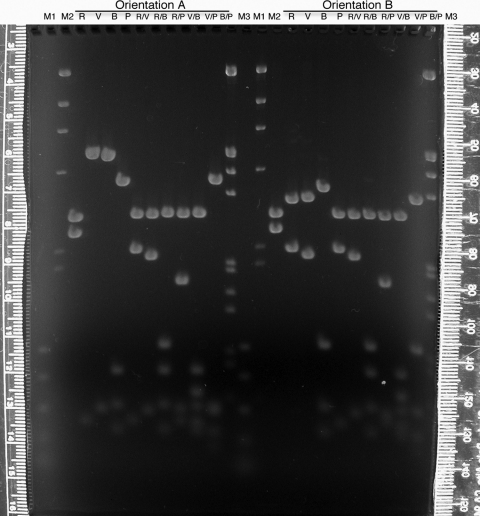
Figure 3:
Mapping gel (1.2% agarose, 1 × TAE) separating restriction enzyme fragments of plasmid DNA containing a 2.6-kb insert of wild-type D. melanogaster DNA from the dusky area of the genome. Students (Fall 2007) used the enzymes EcoRI (R), EcoRV (V), BamHI (B), and PstI (P) individually or in pairs to digest plasmid DNAs containing the insert in both orientations (A or B). After staining with ethidium bromide and destaining in 1 × TAE, the gel was visualized and digitally photographed (Nikon CoolPix995 camera) on an ultraviolet transilluminator (Fotodyne, Hartland, WI). Lambda DNA digested with HindIII (M2) or double digested with HindIII and EcoRI (M3) and a polymerase chain reaction standard (M1; Promega) were used as size markers. The rulers pictured were used to measure migration of the DNA fragments, and each student constructed a standard curve from which the sizes of the DNA fragments were extrapolated. The entire photograph (which is what was placed online for student use) was enhanced (contrast and brightness) using Adobe Photoshop.