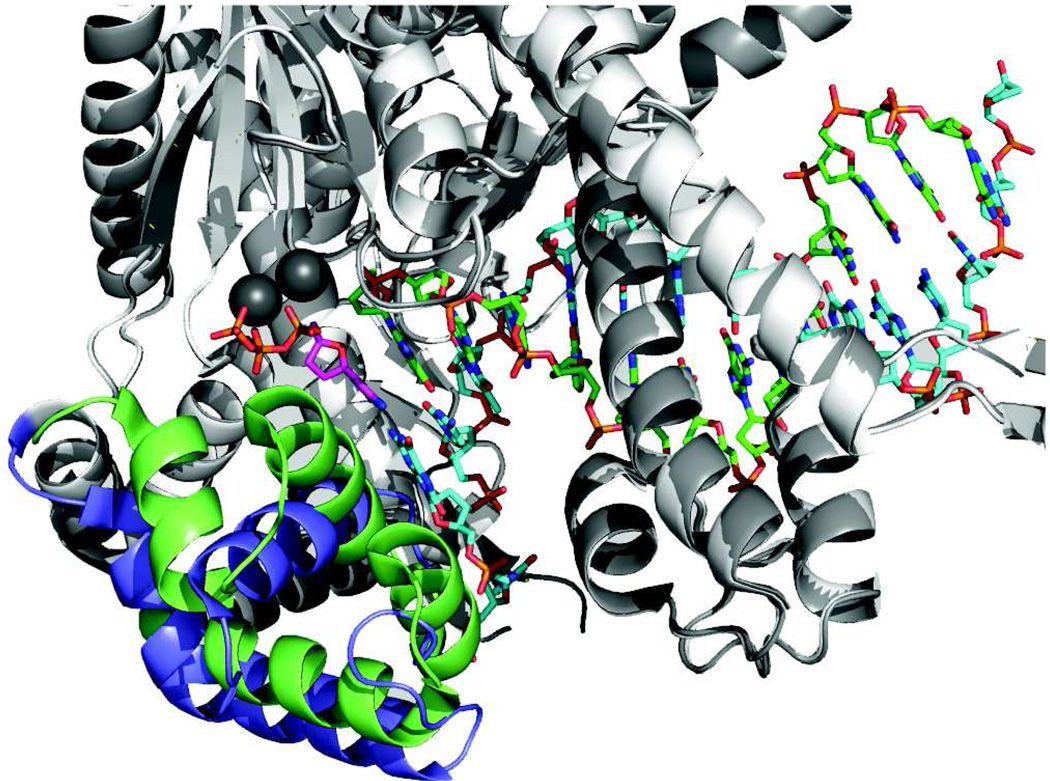
Figure 1. Conformational change upon nucleotide binding.
Structures of T7 DNA polymerase in the presence (green and dark gray) and absence (blue and light gray) of nucleotide are aligned to show the changes in structure of the recognition domain (green or blue). Very little change in structure was seen in the remainder of the protein (light or dark gray), which provided a basis for the alignment. The metal ions are dark gray, the incoming nucleotide is magenta, the primer strand is green and the template strand is cyan. The recognition domain is defined as residues N502 to P560. Drawn using Pymol with PDB files 1tk5 (with nucleotide) and 1tk0 (without nucleotide) [62].