Abstract
The introduction of double stranded RNA (dsRNA) into the cytoplasm of mammalian cells usually leads to a potent antiviral response resulting in the rapid induction of interferon beta (IFNβ). This response can be mediated by a number of dsRNA sensors, including TLR3, MDA5, RIG-I and PKR. We show here that pluripotent human cells (human embryonic stem (hES) cells and induced pluripotent (iPS) cells) do not induce interferon in response to cytoplasmic dsRNA, and we have used a variety of approaches to learn the underlying basis for this phenomenon. Two major cytoplasmic dsRNA sensors, TLR3 and MDA5, are not expressed in hES cells and iPS cells. PKR is expressed in hES cells, but is not activated by transfected dsRNA. In addition, RIG-I is expressed, but fails to respond to dsRNA because its signaling adapter, MITA/STING, is not expressed. Finally, the interferon-inducible RNAse L and oligoadenylate synthetase enzymes are also expressed at very low levels. Upon differentiation of hES cells into trophoblasts, cells acquire the ability to respond to dsRNA and this correlates with a significant induction of expression of TLR3 and its adaptor protein TICAM-1/TRIF. Taken together, our results reveal that the lack of an interferon response may be a general characteristic of pluripotency and that this results from the systematic downregulation of a number of genes involved in cytoplasmic dsRNA signaling.
Key words: dsRNA, interferon, innate immunity, pluripotency, stem cells
Introduction
Cellular responses to dsRNA differ markedly, depending not only on the subcellular location of the duplexes, but also on their size.1 Uninfected mammalian cells rarely express dsRNA within the cytoplasm, most likely because of the ensuing dramatic effects on RNA levels, inhibition of protein synthesis and, if prolonged, cell death (reviewed in ref. 1 and 2). In higher eukaryotes, cytoplasmic dsRNA often triggers the interferon (IFN) signaling pathway and leads to nonspecific inhibition of gene expression. In lower eukaryotes, or in cases where these pathways are lacking or inactive, the RNA interference (RNAi) pathway might provide the primary mechanism to eliminate cytoplasmic dsRNAs (reviewed in ref. 3 and 4). In the nucleus, dsRNAs often serve as substrates for A-to-I editing by members of the ADAR enzyme family, which are ubiquitously expressed in the nuclei of higher eukaryotes.5 Editing can be either site-selective or promiscuous and RNAs edited in these two ways appear to have different functions and fates.1
Multiple complementary and independent systems have been implicated in cytoplasmic dsRNA activity, including PKR (dsRNA-activated protein kinase), the 2–5A system (2′–5′ oligoadenylate synthetase and RNAse L), the helicases MDA5 (melanoma differentiation associated protein 5) and RIG-I (retinoic acid inducible gene I) and the Toll-like receptor 3 (TLR3)-mediated dsRNA response. These systems have been studied in extensive detail in recent years, and there are a number of recent reviews of them (reviewed in refs. 6–14).
PKR is an important player in the cytoplasmic response to dsRNA and induction of IFNβ.15,16 PKR is an IFN-inducible Ser/Thr protein kinase that is directly activated by dsRNA.17 This enzyme is normally present in the cytoplasm in an unphosphorylated, inactive form. Upon activation, a signal transduction cascade is initiated, including the phosphorylation of a number of substrates, such as the eukaryotic initiation factor 2 (eIF-2α)18 and the transcription factor inhibitor IκB.19 Phosphorylation of IκB releases it from the transcription factor NFκB, which can now be translocated to the nucleus where it activates the expression of genes having NFκB binding sites. These genes include IFNβ20 and others.19,21,22
In addition to the PKR pathway, the 2′5′oligoadenylate synthetase (2′5′-AS)/RNAse L pathway responds to dsRNA. 2′5′-AS is upregulated in IFN-treated and virus-infected cells23 and is activated upon binding to dsRNA.2,24 RNAse L, a widely expressed cytoplasmic endoribonuclease, dimerizes and is activated by 2′5′AS.25 Activated RNAse L catalyzes the degradation of viral and cellular RNAs,26 thus inhibiting protein synthesis.27
Other cytoplasmic dsRNA-activated signaling pathways have recently been shown to be even more important for the induction of IFNβ. TLR3 is localized to endosomes in myeloid dendritic cells28,29 but is found on both the cell surface30 and endosomes28 of fibroblasts and epithelial cells. TLR3 signaling is initiated by binding to dsRNA and results in the recruitment of cytoplasmic adapter proteins that contain TIR domains.6 A key target for dsRNA-mediated TLR3 signaling is TICAM-1/TRIF.31 TRIF is recruited to endosomes following dsRNA recognition by TLR3 and functions as a docking platform that interacts with several signaling proteins to initiate divergent signaling pathways leading to activation of IRF3, NFκB and the IFNβ promoter.7,31–38 Yet other important pathways involve the recognition of dsRNA by the cytoplasmic RNA helicases RIG-I and MDA5 (reviewed in ref. 13 and 39). RIG-I recognizes single-stranded RNAs with 5′-phosphates,40 RNAs with 5′-triphosphates41 and dsRNAs of intermediate length.42 Signaling by RIG-I is affected by a number of factors,13,43 including the important RIG-I adaptor MITA (mediator of IRF3 activation)44 or STING (stimulator of interferon genes).45 On the other hand, MDA5 preferentially responds to long dsRNAs.42 Like TLR3, signaling for both RIG-I and MDA5 proceeds through NFκB and IRF3.13,39 Upon binding to RNAs, RIG-I and MDA5 associate with the outer mitochondrial membrane associated adapter protein IPS-1 (IFNβ promoter stimulator-1), also known as MAVS (mitochondrial antiviral signaling), VISA (virus-induced signaling adapter) or CARDIF (CARD adapter inducing IFNβ),46–49 which plays an essential role in RIG-I/MDA5 signaling.50,51 Finally, it has recently been reported that EYA4 (Eyes absent 4) associates with IPS-1 and STING and can stimulate the expression of IFNβ in response to the undigested DNA of apoptotic cells and enhances the innate immune response against viruses.52
The precise mechanisms by which dsRNA enters cells and is delivered to cytosolic sensors are still unknown. Several studies have followed dsRNA uptake into cells and interaction with the TLR3 pathway.29,35,53,54 Johnsen et al.53 examined the dynamics of dsRNA internalization using fluorescent RNA. In the absence of transfection reagents these authors saw uptake into early endosomes (consistent with clathrin-mediated uptake), followed by eventual transit to lysosomes. In these cells, dsRNA colocalized with endosomal TLR3. Further, introduction of dsRNA into cells via transfection may lead to a similar outcome. Various lipofection agents deliver RNAs to endosomes,35 and dsRNA transfection was seen to lead to TRIF recruitment to endosomes within 20 minutes, leading to the appearance of “speckles” in the cytoplasm.54
In this study, we examined the cytoplasmic responses to dsRNA in undifferentiated and differentiated hESCs and in induced pluripotent stem (iPS) cells. Following transfections with dsRNA, we found that while differentiated cells activate interferon expression, hES and iPS cells do not. In addition, key factors that are involved in the cytoplasmic responses to dsRNA have been carefully examined, yielding important clues into the underlying basis for this phenomenon.
Results
hESCs have a weak IFNβ response to dsRNA.
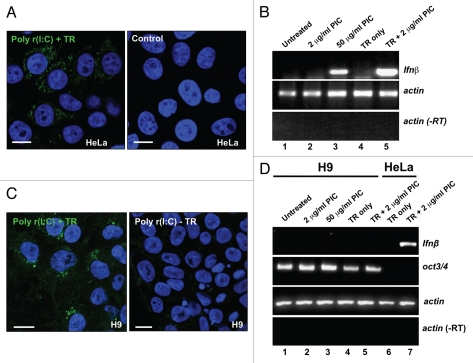
We first asked whether hESCs can respond to dsRNA by activating the expression of IFNβ. HeLa and H9 cells were treated with poly r(I:C) (PIC; polyriboinosinic:polyribocytidylic acid, a synthetic analog of viral dsRNA) either in the presence or absence of a transfection reagent. Figure 1A shows that 6 hrs after transfection of HeLa cells with PIC, dsRNA is readily detected in the cytoplasm in a punctate distribution consistent with previous reports that dsRNA uptake is into endosomes.53,54 A similar pattern was observed after adding PIC directly to HeLa cells (Suppl. Fig. 1). Figure 1B shows that such treatments lead to the strong and rapid induction of transcription of IFNβ in HeLa cells (Fig. 1B, lane 5). IFNβ induction in the absence of transfection was only seen with high concentrations of PIC (Fig. 1B, lanes 2 and 3 and Suppl. Fig. 1). While PIC could be efficiently introduced to hESCs via transfection, dsRNA uptake was never observed in these cells without transfection reagents (Fig. 1C and Suppl. Fig. 1), suggesting that active dsRNA internalization in hESCs differs from that in HeLa cells. More interestingly, and in contrast to results seen with HeLa cells, IFNβ was not induced by dsRNA in hES cells, regardless of whether they were grown under feeder-free conditions (Fig. 1D and data not shown) or on irr-MEFs (Suppl. Fig. 2). Since 12d old irr-MEFs still retain a weak IFNβ response (Suppl. Fig. 2), we examined the IFNβ response in hESCs and iPS cells under feeder free conditions throughout this study. The IFNβ response in another hES cell line (H14) was also almost undetectable (Suppl. Fig. 3).
Figure 1.
dsRNA does not induce IFNβ in hES cells. (A) PIC was delivered into HeLa cells via transfection. Left, 6 hrs after transfection of PIC, HeLa cells were fixed and incubated with monoclonal antibody J2 which specially recognizes long dsRNAs. Right, HeLa cells without dsRNA treatment. (B) A rapid and strong IFNβ response in HeLa cells. Lane 1, no PIC treatment; lane 2, 2 µg/mL PIC was added to growth media for 6 hrs; lane 3, 50 µg/mL PIC was added to growth media for 6 hrs; lane 4, treatment of cells with transfection reagent (TR) Lipofectamine 2000 (LF) alone; lane 5: 2 µg/mL PIC was transfected into cells with LF for 6 hrs. Actin was used as a loading control and -RT was used as a RT quality control. (C) PIC was delivered into H9 cells by transfection with Fugene HD. Left, H9 cells were cultured on Matrigel and were transfected with 2 µg/mL PIC for 6 hrs. Right, 50 µg/mL PIC was added to medium of H9 cells for 6 hrs. (D) No detectable IFNβ response in H9 cells after 6 hrs PIC treatment. Lane 1, no PIC treatment; lane 2, 2 µg/mL PIC was added to growth media for 6 hrs; lane 3, 50 µg/mL PIC was added to growth media for 6 hrs; lane 4, H9 cells were treated with Fugene HD alone; lane 5: H9 cells were transfected with 2 µg/mL PIC for 6 hrs; lane 6, HeLa cells were treated with Fugene HD alone; lane 7: HeLa cells were transfected with 2 µg/mL PIC for 6 hr using Fugene HD. Oct3/4 is a stem cell pluripotency marker. Actin was used as a loading control and -RT was used as a RT quality control.
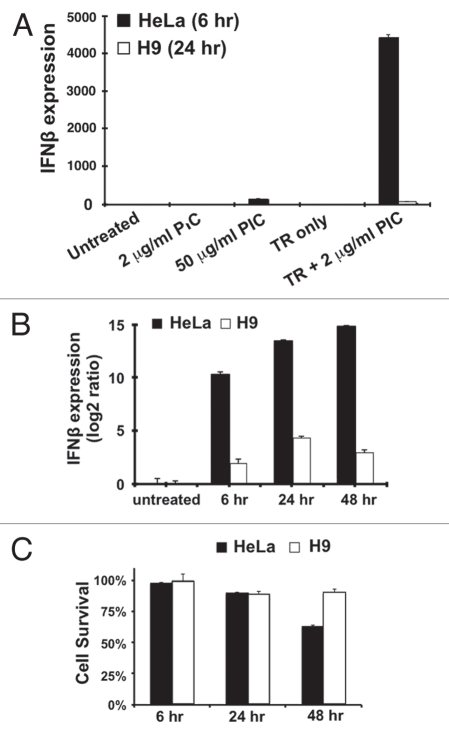
We next set up a more quantitative analysis of these responses by using real-time RT-qPCR (Fig. 2). While in HeLa cells there is a dramatic induction of IFNβ mRNA within 6 hrs, the signal in H9 cells is barely detectable (Fig. 2A and B). Furthermore, the induction of IFNβ mRNA in HeLa cells was even more dramatic after prolonged treatment; however, the barely detectable IFNβ signal in H9 cells remained almost unchanged after 48 hrs post-transfection with PIC (Fig. 2B). Since the response was several orders of magnitude weaker than that observed in HeLa cells, we speculate that this extremely weak IFNβ signal in H9 cells may result either from a very weak intrinsic dsRNA response system or from a small fraction of cells in our hESC cultures that have begun to differentiate. In addition, the lack of an IFNβ response in H9 cells is not due to loss of cell viability (Fig. 2C).
Figure 2.
Attenuated dsRNA response in hES cells. (A) The IFNβ responses in HeLa and H9 cells were quantitatively measured by RT-QPCR and normalized to each endogenous actin mRNA. The treatments were described as in Fig. 1B. Note that the IFNβ response was almost undetectable in H9 cells after the PIC treatment. (B) The IFNβ responses in HeLa and H9 cells during a time-course treatment. 2 µg/mL PIC was transfected into both cell lines and IFNβ mRNA was measured 6 hrs, 24 hrs and 48 hrs post-transfection. (C) Cell survival rates after the PIC transfection. All cells were disassociated from plates at various time points and viable cells were counted using trypan blue. The cell survival rate was calculated using the formula: (# uncolored cells)/(# uncolored cells + # blue cells).
Expression of genes involved in cytoplasmic responses to dsRNA in hESCs.
As a first step towards a molecular understanding of how pluripotent cells respond to dsRNAs, we used a genome-wide approach to determine the expression pattern of a number of the key genes involved. We therefore isolated cytoplasmic polyadenylated RNAs from both HeLa and H9 cells and subjected them to high throughput sequencing using the Illumina/Solexa platform. Equivalent numbers of 75-nucleotide sequence reads were aligned to the genome using the UCSC Genome Browser and the number of reads aligning to annotated exons of genes were summed. Some of the key results obtained are shown in Table 1 and Table S1. The numbers shown represent the normalized relative levels of mRNA expression between HeLa and H9 cells. Consistent with our previous studies,55 Gapdh and Adar1 are each expressed at similar levels in H9 cells and in HeLa cells. The stem cell markers Nanog, Sox2, Lin28 and Oct3/4 are all highly expressed in H9 cells but absent in HeLa cells.
Table 1.
Relative mRNA quantitations
| Gene | RPKM HeLa | RPKM H9 | Function |
| Gapdh | 1230 | 1727 | Internal control |
| Nanog | 0.05 | 21.8 | Stem cell marker |
| Sox2 | 0.00 | 70.6 | Stem cell marker |
| Lin28 | 0.03 | 285.0 | Stem cell marker |
| Oct3/4 | 0.2 | 560 | Stem cell marker |
| TLR3 | 1.3 | 0.1 | IFNβ induction |
| TICAM-1/TRIF | 11.8 | 0.4 | IFNβ induction |
| RIG-I | 0.8 | 1.0 | IFNβ induction |
| STING/MITA | 18.1 | 0.1 | IFNβ induction |
| TBK1 | 6.0 | 2.0 | IFNβ induction |
| IKBKE | 2.0 | 3.3 | IFNβ induction |
| MDA5 | 1.9 | 0.2 | IFNβ induction |
| VISA/IPS-1/MAVS | 6.9 | 6.8 | IFNβ induction |
| EYA4 | 48.6 | 0.0 | IFNβ induction |
| PKR | 14.1 | 4.1 | IFNβ induction |
| NFκB1 | 8.3 | 1.8 | IFNβ induction |
| NFκB2 | 16.2 | 6.3 | IFNβ induction |
| IκB | 6.3 | 4.6 | IFNβ induction |
| IRF3 | 10.9 | 19.3 | IFNβ induction |
| IRF7 | 0.4 | 2.6 | IFNβ induction |
| OAS1 | 8.7 | 0.05 | OAS, RNAseL pathway |
| OAS3 | 12.7 | 0.4 | OAS, RNAseL pathway |
| RNAse L | 0.68 | 0.1 | OAS, RNAseL pathway |
| ADAR1 | 58.3 | 53.5 | Editing (nuclear) |
Relative expression of genes involved in the cytoplasmic pathways to dsRNA between HeLa and H9 cells. Equal amounts of poly(A)+ RNAs were analyzed using the Illumina Genome Analyzer. The relative levels of mRNA expression are reported as the normalized number of reads that mapped to the annotated exons of each gene (# of reads per thousand nucleotides of exons, per million total reads [RPKM]). Higher numbers are positively correlated to higher levels of gene expression. Reads from one experiment are reported, but similar relative levels of mRNA expression were seen in two other independent sequencing experiments.
The deep sequencing work offered a number of important clues to the underlying dsRNA sensing defects in hES cells. First, H9 cells exhibit a severe defect in the expression of MDA5, TLR3 and its key signaling adapter, TRIF, which are crucial factors involved in IFNβ induction (Table 1). Second, although RIG-I is expressed in pluripotent cells, we found a number of its positive regulators are expressed at lower levels in H9 cells than those in HeLa cells (Table 1). In contrast, a number of its negative regulators are expressed at higher levels in H9 cells (Table S1). Among these factors, the recently identified MITA/STING, which has been shown to be very important for RIG-I mediated signaling,44,45 is completely absent in H9 cells, as is EYA4, which has recently been shown to be important for innate immunity and to interact with IPS-1 and MITA52 (Table 1). Although the role of RIG-I regulators and the nature of RNA ligands for RIG-I remain somewhat controversial,56 the possibility exists that one or more regulators of RIG-I might function together to attenuate PIC-stimulated IFNβ signaling via RIG-I in hESCs. Third, deep sequencing revealed that some of the key downstream signaling targets of PKR (IκB, NFκB and IRF3) are abundantly expressed in H9 cells (Table 1), suggesting that the failure to activate the PKR signaling pathway is not the result of low expression levels of some of its key factors. Finally, RNAse L mRNA levels are low in H9 cells compared to HeLa cells and none of the known forms of oligoadenylate synthetases (OAS enzymes) are expressed at a significant level in H9 cells (Table 1, and data not shown). This means that the RNAse L pathway cannot be activated directly in these cells.
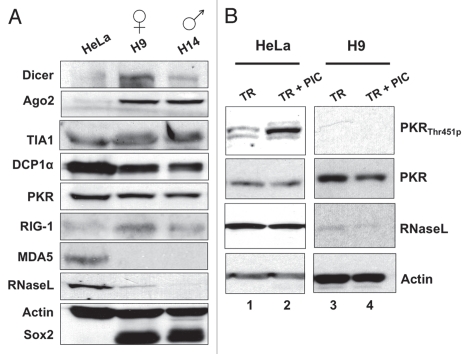
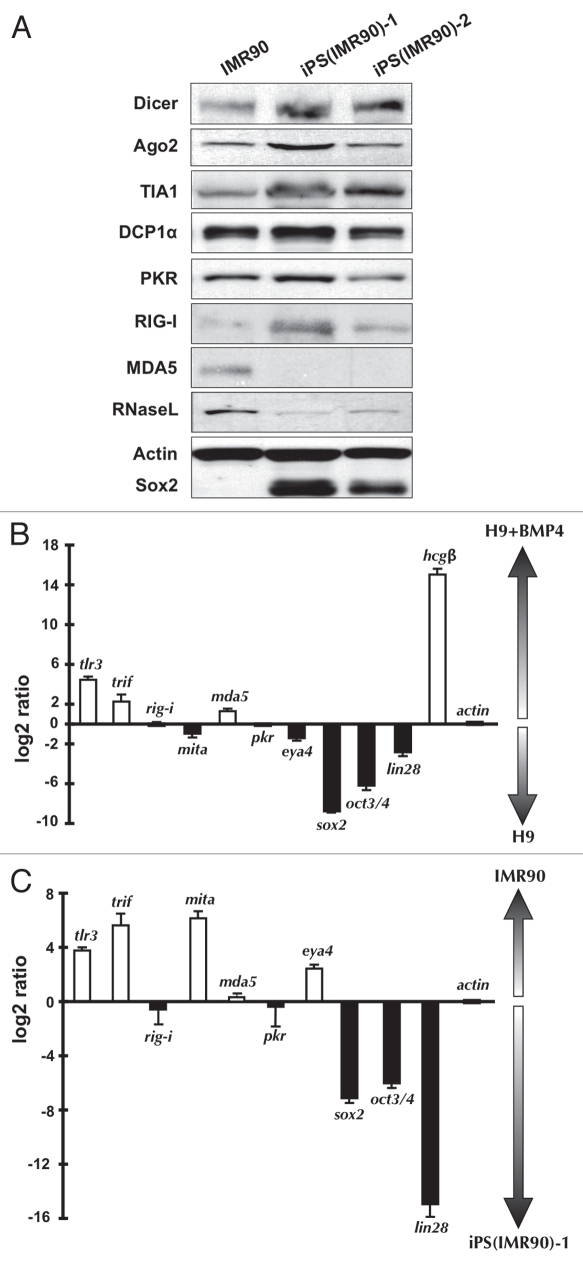
Western blotting of extracts prepared from two different hESC lines, H9 and H14, confirmed what we had observed by deep sequencing (Fig. 3A). H9 and H14 both express Dicer and Ago2, which are important for RNAi and microRNA responses. Such responses are well documented to be robust in pluripotent cells.57–61 In addition, markers of cytoplasmic stress granules (TIA1) and processing bodies (P-bodies) (DCP1α) are also expressed well. However, several sensors involved in dsRNA-triggered IFNβ signaling are altered dramatically in their expression. Consistent with the deep sequencing results, hESCs and iPS cells express PKR and RIG-I at similar levels to those seen in HeLa and IMR90 cells, but RNase L levels are much lower and MDA5 is undetectable (Fig. 3A). Numerous attempts to detect TLR3 in these cells were unsuccessful (data not shown).
Figure 3.
Some factors involved in cytoplasmic responses to long dsRNAs are expressed in hESCs. (A) Whole cell extracts were collected from HeLa cells and hESCs grown under feeder-free conditions and equal amounts of total proteins were subjected to SDS-PAGE and analyzed by western blotting with the indicated antibodies. Actin was used as a loading control and Sox2 was used as a marker for pluripotent cells. (B) Lack of dsRNA-induced PKR autophosphorylation in H9 cells. HeLa cells and H9 cells were transfected with 2 µg/mL PIC for 6 hrs and 24 hrs respectively, and whole cell extracts were collected from the untreated cells and the PIC treated cells at the indicated time points. Equal amounts of total proteins were loaded. Actin was used as a loading control.
The above results offered strong support for some of the observed defects in dsRNA signaling observed in hESCs. However, they did not allow us to understand the failure of IFNβ activation by dsRNAs via PKR. In fact, this activation is accompanied by autophosphorylation at Thr451.62,63 Using antibodies specific for this modification, we observed a strong increase in phosphorylation following transfection with dsRNA (Fig. 3B, lane 2). However, autophosphorylation was undetectable in H9 cells (Fig. 3B, lane 3), even after the transfection of PIC (Fig. 3B, lane 4). We do not yet know the molecular basis for this activation defect, but hypothesize that it may underlie at least some of the absence of a vigorous IFN response in pluripotent cells.
The lack of a cytoplasmic response to dsRNA correlates with pluripotency.
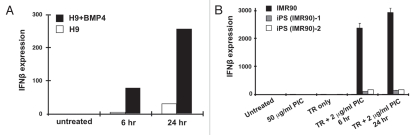
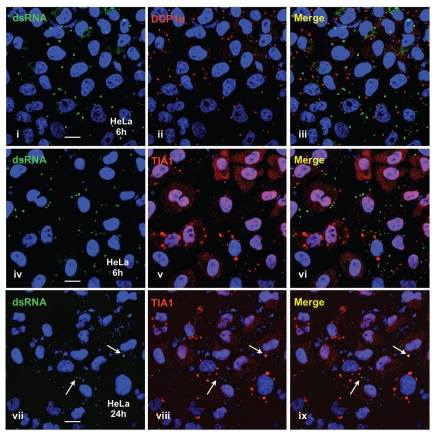
The above results suggest that weak dsRNA signaling may be an intrinsic characteristic of pluripotency. In order to address this we performed several complementary experiments. First, we showed that the poor dsRNA response in H9 cells is not due to an irreversible defect. We treated H9 cells with BMP4 and then examined the cultures for IFNβ induction. BMP4 induces the differentiation of hES cells into trophoblasts.64,65 After a short period of differentiation (4 days), we observed strongly enhanced (about 10-fold) IFNβ induction (Fig. 4A). This level of induction is still about 10-fold lower than that seen in HeLa cells (compare Fig. 2A and B with Fig. 4A). Second, we examined pluripotent cells that have been derived from differentiated cells (iPS cells). The iPS cells we used for this study were derived from human embryonic fibroblast IMR90 cells.66 Figure 4B shows that the iPS precursor cells, IMR90 cells, show a robust IFNβ response after transfection with PIC. By using a similar transfection method, we were able to introduce PIC into two independent iPS cells with a similar transfection efficiency to IMR90 and hESC lines (as shown by equivalent numbers of cytoplasmic foci formed by dsRNAs; Suppl. Figs. 4 and 5). However, each iPS line exhibited a 20–30 fold lower IFNβ response, suggesting that reprogramming leads to the loss of dsRNA response in the cytoplasm.
Figure 4.
A weak cytoplasmic dsRNA response is a general feature of pluripotency. (A) Differentiated hESCs gain an IFNβ response to PIC. H9 cells and BMP4-treated H9 cells were transfected with 2 µg/mL PIC for 6 hrs or 24 hrs. IFNβ responses were quantitatively measured by RT-QPCR and normalized to each endogenous actin mRNA. Experiments were at least duplicated and error bars were calculated from three biological repeats. (B) Reprogrammed iPS cells lack an IFNβ response to PIC. iPS (IMR90) cells were treated with either 50 µg/mL PIC directly, or transfected with 2 µg/mL PIC for 6 hrs or 24 hrs. The IFNβ responses were quantitatively measured by RT-QPCR and normalized to each endogenous actin mRNA.
Western blotting confirmed that IMR90 cells closely resemble HeLa cells in the expression of some key cytoplasmic dsRNA sensors (compare Figs. 3A and 5A). Further, two independent iPS lines generated from IMR90 cells are indistinguishable from H9 and H14 cells in their patterns of expression of these same factors (compare Figs. 3A and 5A). Taken together, the above results lead us to conclude that a strongly attenuated dsRNA response is a general feature of pluripotent human cells, and that this attenuated response is associated with the lack of expression of a specific set of proteins.
Figure 5.
Factors involved in cytoplasmic responses to long dsRNAs in iPS cells and following differentiation of hESCs to trophoblasts. (A) Whole cell extracts were collected from IMR90 cells and iPS cells grown under feeder-free conditions and equal amounts of total proteins were subjected to SDS-PAGE and analyzed by western blotting with the indicated antibodies. Actin was used as a loading control and Sox2 was used as a marker for pluripotent cells. (B) Real-time RT-PCR was used to compare the relative levels of the mRNAs for the indicated factors between H9 cells and H9 cells that had been treated with BMP4 to promote differentiation to trophoblasts. Note that the bars represent the relative ratios of mRNA concentrations between the two cell populations (normalized to actin expression), not the absolute levels of transcripts. White bars, transcripts preferentially expressed in trophoblasts. Black bars, transcripts preferentially expressed in H9 cells. sox2, oct3/4 and lin28, pluripotency markers. hcgβ, trophoblast marker. (C) Real-time RT-PCR was used to compare the relative levels of the mRNAs for the indicated factors between IMR90 cells and iPS cells derived from IMR90 cells. Bars are as in (B). Another iPS cell line, iPS(IMR90)-2 showed essentially identical results (data not shown.)
The TLR3/TRIF pathway correlates with differentiation-mediated IFNβ response to dsRNA.
To further explore which dsRNA response pathway is induced upon differentiation, we used real-time quantitative RT-PCR to compare the levels of expression of many key factors during differentiation of H9 cells into trophoblasts (Fig. 5B). Results showed that TLR3 and TRIF were significantly upregulated upon differentiation, while other signaling factors remained essentially unchanged. As these were the only factors strongly upregulated, we speculate that this underlies our findings that the IFNβ response in these cells is less robust than that in HeLa cells, where more factors are expressed. These observations were further supported by examination of changes associated with reprogramming of IMR90 cells to iPS cells (Fig. 5C). Here, reprogramming led to a strong downregulation of both TLR3 and TRIF, although in this case MITA and EYA4 (involved in the RIG-I pathway) were also significantly reduced. These results are generally in excellent agreement with those obtained by western blotting and deep sequencing. Taken together, these results point to the likely importance of TLR3 and TRIF in the acquisition of a dsRNA response in trophoblasts and the repression of TLR3, TRIF, MITA and EYA4 upon reprogramming of IMR90 cells.
dsRNA uptake and fate in hESCs and iPS cells.
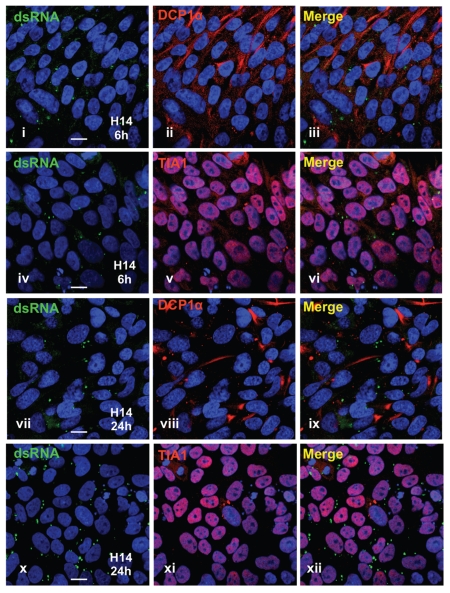
The above results demonstrate that hESCs and iPS cells do not induce IFNβ in response to dsRNA. While much or most of this defect likely results from the low expression profiles of TLR3, TRIF, MITA and MDA5 (Table 1) and the inability of dsRNA to activate PKR (Fig. 3B), we could not rule out the additional possibility that hESCs and iPS cells retain some ability to respond to dsRNA, but do not internalize dsRNAs efficiently. Activation of both PKR and RIG-I by dsRNA requires that the dsRNA be present in the cytoplasm. We therefore carried out a number of studies to investigate the cellular uptake and fate of dsRNA in HeLa cells, hES cells and iPS cells. Figure 6 shows the results from some of these studies in HeLa cells. Using an antibody specific for dsRNA longer than 40 bp in length,67 we observed that when transfected or directly delivered into HeLa cells, PIC initially accumulates primarily in discrete cytoplasmic foci (Figs. 1A and 6i, 6iv and Suppl. Fig. 4). These foci are not processing bodies (P-bodies; reviewed in ref. 68 and 69), as indicated by lack of colocalization with DCP1α (Fig. 6ii and 6iii) or stress granules (reviewed in refs. 68 and 69), as indicated by lack of colocalization with TIA1 (Fig. 6v and 6vi). They likely represent endosomal compartments reported previously as the sites of dsRNA uptake.35,54 Interestingly, at 24 hrs after transfection, the vast majority of the dsRNA disappeared from the transfected cells. By this time the cells have mounted a vigorous response involving IFN induction. In HeLa cells dsRNA leads to the rapid appearance of stress granules (Fig. 6v). Significantly, the small amount of dsRNA that is still detectable in HeLa cells at 24 hrs colocalizes with stress granules (Fig. 6vii–ix, arrows).
Figure 6.
Visualization of transfected dsRNAs in HeLa cells. HeLa cells were transfected with 2 µg/mL PIC for 6 hrs and 24 hrs, and then stained with anti-dsRNA J2 antibody and anti-DCP1α (i–iii) or J2 and anti-TIA1 (iv–ix). Note that dsRNAs do not colocalize with DCP1α or TIA1 at 6 hrs post-transfection. dsRNA-associated cytoplasmic dots are largely diminished after 24 hrs in cells, and the remaining dots colocalize with TIA1-positive stress granules (arrows in vii–ix).
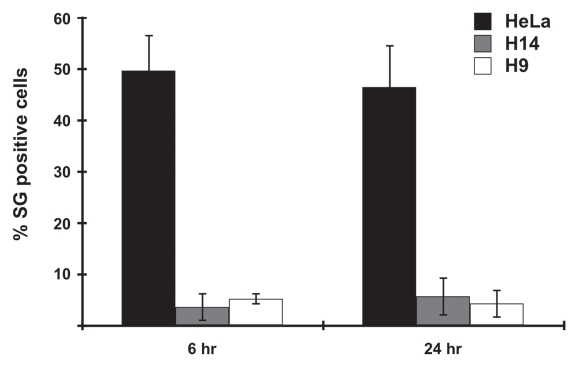
Pluripotent cells exhibit several important differences in this type of analysis. Transfection of H14 cells with PIC leads to cytoplasmic dots that are visibly indistinguishable from those seen in HeLa cells (Fig. 7). This is also observed for H9 cells and iPS cells (Suppl. Figs. 4 and 5). Also as in HeLa cells, dsRNA in ES cells does not colocalize with DCP1a or TIA1 (Fig. 7iii and vi). Interestingly, however, in hES cells DCP1a is broadly distributed throughout the cytoplasm (for e.g., see Fig. 7ii), in contrast to the situation in HeLa cells where distinct cytoplasmic processing bodies (P-bodies) are observed (Fig. 7ii). Also and in stark contrast to the observations in HeLa cells, cytoplasmic dsRNA in hES cells (Fig. 7vii and x) or iPS cells (Suppl. Fig. 5) is still readily detected in cytoplasmic foci at 24 hrs after transfection. We have observed virtually no colocalization of dsRNA with stress granules in these cells, and note that many fewer stress granules are induced in pluripotent cells after the same transfection treatment (Fig. 8). Taken together, these localization studies may shed new light on the inability of pluripotent cells to express IFNβ in response to transfected dsRNA.
Figure 7.
Visualization of transfected dsRNAs in human ES cells. H14 cells were transfected with 2 µg/mL PIC for 6 hrs and 24 hrs, and then fixed and stained with anti-dsRNA J2 antibody and anti-DCP1α (i–iii and vii–ix) or J2 and anti-TIA1 (iv–vi and x–xii). Note that P-bodies (indicated by DCP1α) are larger and more broadly distributed in hESCs; less visible stress granules (indicated by TIA1) are visible in hESCs (also see Fig. 8); however, dsRNAs remain detectable in cytoplasmic dots and do not colocalize with DCP1α or TIA1 24 hrs post-transfection.
Figure 8.
Fewer visible stress granules (SGs) are formed in hES cells following transfection stress. HeLa, H9 and H14 cells were transfected with 2 µg/mL PIC for 6 hrs and 24 hrs respectively, and then stained with J2 antibody and anti-TIA1. SG-positive cells were counted under 3–6 randomly selected different microscope fields, each containing 100–200 cells. Standard deviation was calculated from all counted fields in three independent experiments.
Discussion
Most mammalian cells respond vigorously to cytoplasmic dsRNA through the activation of PKR and RNAse L as well as through signaling cascades mediated by TLR3/TRIF, RIG-I and MDA5. These potent response pathways play major roles in cellular antiviral responses and complement (and in fact largely supplant) the more ancient innate immunity to dsRNA provided by the RNAi machinery. We have shown here that hES cells do not induce interferon in response to dsRNA and that this defect is the result of several important differences between pluripotent cells and differentiated cells (Fig. 9). First, although PKR and at least several important downstream targets are expressed in hESCs, they are not activated by dsRNA, because we see no evidence of PKR autophosphorylation and IFNβ is not induced (Figs. 2–3). Second, the amounts of TLR3, the TLR3 adaptor TRIF, the RIG-I adaptor MITA and the helicase MDA5 are greatly diminished (Figs. 3, 5 and Table 1). Finally, RNAse L and OAS are expressed at very low or negligible levels (Figs. 3, 5 and Table 1). Together, these defects likely lead to an inability of stem cells to respond through any of the currently known cytoplasmic dsRNA-mediated sensing and signaling pathways.
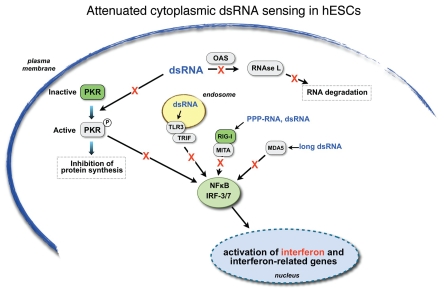
Figure 9.
Attenuated cytoplasmic dsRNA sensing in hESCs. The major cytoplasmic dsRNA signaling pathways are shown. TLR3 is shown as associated with endosomes, while in some cells it may also appear on the plasma membrane. See text for details. Proteins depicted by green boxes are expressed in hES cells, while those depicted by grey boxes are not. The red “X”s indicate that the signaling pathway for each known dsRNA response system is severely compromised in pluripotent cells.
While signaling pathways initiated by dsRNA delivered by transfection are clearly compromised, we do not yet know whether pluripotent cells completely lack the ability to mount an immediate innate immune response to viral infections. RIG-I is important for IFNβ responses to a variety of ssRNA viruses carrying either a plus-strand RNA genome (e.g., hepatitis C virus, Japanese encephalitis virus) or a minus-strand RNA genome (e.g., influenza virus, Sendai virus, vesicular stomatitis virus, measles virus, respiratory syncytial virus and Ebola virus), whereas encephalomyocarditis virus and Theiler's encephalomyelitis virus induce IFNβ through the MDA5 pathway.70,71 Since RIG-I is still expressed in them, it will be of interest in future studies to infect pluripotent cells with viruses that are known to induce IFNβ through the RIG-I pathway.
One puzzling aspect of the above results is that in hES cells PKR is expressed but not activated by exogenous dsRNA. A possible explanation for this observation is that the PKR enzyme is altered in some way in pluripotent cells such that it cannot be activated by dsRNA. We have no evidence to support this model but cannot rule it out at this time. Our data, however, suggest an alternative model that dsRNA delivered to these cells by transfection is not allowed access to cytoplasmic PKR and perhaps also to other sensors such as RIG-I. Immunofluorescence studies show that dsRNA appears to remain sequestered in cytoplasmic foci after transfection of hES cells or iPS cells (Figs. 1 and 6, 7; Figs. S1, S4 and S5), while in HeLa cells it disappears from these sites within 24 hours (Fig. 6). In order to access PKR and induce IFN, at least some of the transfected dsRNA in HeLa cells must be released into the cytoplasm. This may not occur in pluripotent cells.
Why do hESCs differ so dramatically from differentiated cells in their cytoplasmic response to dsRNA? We suggest several possible reasons for this. First, pluripotent cells may lack a dsRNA-inducible IFN response pathway because this represents an antiviral response pathway that is not needed in them. Viral infection of these cells might seriously jeopardize their ability to maintain their ability to differentiate or to maintain effective pluripotency. Therefore, it may be better for the organism for these cells to die rather than to risk trying to fight infections. A second possibility is that one or more components of the IFN signaling pathway is incompatible with pluripotency or differentiation. We do not yet know how expression of any of the repressed factors described here affects hESC biology but do note that differentiation into trophoblasts leads to the induction of TLR3 and TRIF (Fig. 5B). Third, pluripotent cells may lack this pathway because in these cells some important dsRNAs or highly structured RNAs may be exported from the nucleus to the cytoplasm. In differentiated cells dsRNAs are rarely present in the cytoplasm, but are frequently formed in the nucleus as the result of sense-antisense transcription or inverted repeats within transcripts. Such duplexes can serve as targets for promiscuous ADAR-mediated editing, which leads to nuclear retention.72 We have recently shown that while nuclear editing in hES cells is robust, the nuclear retention mechanism is not.55 Thus, mRNAs or dsRNAs that would normally be nuclear retained in differentiated cells might be exported to the cytoplasm in pluripotent cells. Some of these might have the potential to activate PKR or RIG-I. If the IFN response system were operable, these might seriously impact cell growth or viability. However, in hESCs cytoplasmic dsRNAs may be able to access the RNAi pathway, leading to the generation of a pool of “endogenous siRNAs” that are not found in differentiated cells. These could serve an important biological purpose, such as transposon silencing, which is accomplished in germ cells via the alternative piRNA pathway.73 Consistent with this, there are several recent reports of endogenous, repeat-derived small RNAs in embryonic mouse cells.74–76 Finally, it has recently been shown that components of the endogenous small RNA processing machinery may have important functions in the maintenance of stem cell properties.77–79
An attenuated cytoplasmic response to dsRNA may be a general feature of pluripotency. In undifferentiated EC cells and in normal embryonic cells, IFN genes are not induced by dsRNA.80–83 The expression of both interferon regulatory factor (IRF) and IFN genes is developmentally regulated in mouse EC cells; these genes become functional only after cell differentiation.83 Likewise, cellular levels of RNase L are regulated during differentiation of murine embryonal carcinoma cells.84 It has been suggested that the key defect in this response is the lack of activation of the proper transcription factors for these genes.85 While dsRNA does not induce IFN in F9 cells, these cells do respond to dsRNA after differentiation into cells of parietal endoderm lineage.82
Finally, what are the implications of our findings for dsRNA-mediated gene silencing in hES cells? The lack of an interferon response in pluripotent cells suggests that it might be possible in these cells to use long dsRNA instead of shorter (and more expensive) siRNAs to induce specific gene expression knockdown in stem cells. In fact, specific dsRNA-induced RNAi silencing has been reported in mouse ES cells.60,61 However, in these studies the extent of knockdown was quite modest. Our results lead us to suggest that it may generally prove to be difficult to use long dsRNAs for efficient gene silencing in pluripotent cells. Since transfected dsRNAs may be retained in endosomes in hES cells, they may not only be unavailable to activate PKR and RIG-I, but also may serve as poor substrates for cytoplasmic Dicer enzyme.
Materials and Methods
Cells, differentiation and dsRNA transfection.
HeLa cells were cultured under standard conditions. Passages 2 to 6 of IMR90 cells were used in this study. Human iPS (IMR90) cell lines were generated from IMR90 precursor cells and were verified at the UConn Stem Cell Core86 and were confirmed positive for Tra-1-81, Tra-1-60, SSEA-3 and SSEA-4 by immunofluorescence and teratoma formation.66 hES H9, H14 cell lines and iPS cell lines were maintained on plates coated with Matrigel (BD Biosciences) in either defined mTeSR medium (StemCell Technologies Inc.,) or conditioned medium (CM) with irradiated mouse embryo fibroblasts supplemented with 4 ng/mL human Basic Fibroblast Growth Factor (bFGF) (Life Technologies).55 Cell cultures were regularly evaluated for Oct3/4 expression every 3–4 weeks and cells were passaged every 6–7 days. For trophoblast differentiation, hES cells were treated with 100 ng/mL BMP4 (R&D Systems) in the presence of CM and bFGF for 4–6 days.64,65
Transfection of dsRNAs used Lipofectamine 2000 (Invitrogen) or Fugene HD (Roche) according to the manufacturer's protocol. Transfection of dsRNA into H9, H14 and iPS cells was carried out at day 2 or day 3 post-passage using Fugene HD (Roche).
Immunofluorescence microscopy.
Cells were fixed in 4.0 % paraformaldehyde for 20 min at RT, then permeabilized in PBS containing 0.2–0.5% Triton X-100 on ice for 5 min. The detergent concentration varied depending on the cell line used. The following primary antibodies were used for the colocalization studies: mouse anti-dsRNA, J2 antibody (1:200, English & Scientific Consulting, Hungary), rabbit anti-DCP1α (1:200, Sigma) and goat anti-TIA1 (1:50, Santa Cruz Biotechnology). Suitable fluorescence conjugated secondary antibodies were used to detect each specific primary antibody. The nuclei were counterstained with TOPRO-3. Images were taken with a Zeiss LSM 510 microscope.
Immunoblots.
Whole cell lysates from HeLa, IMR90, hES and iPS cells were collected and resolved on SDS-PAGE gel. Primary antibodies used in this study were rabbit anti-Sox2 (1:1,000, Millipore), mouse anti-RNase L (E-9) (1:1,000, Santa Cruz Biotechnology), rabbit anti-PKR (K-17) (1:1,000, Santa Cruz Biotechnology), rabbit anti-p-PKR (Thr451) (1:200, Santa Cruz Biotechnology), rabbit anti-Ago2 (1:500, Cell Signaling), rabbit anti-Dicer (1:200, Cell Signaling), rabbit anti-DCP1α (1:1,000, Sigma), goat anti-TIA1 (1:500, Santa Cruz Biotechnology), rabbit anti-RIG-I (1:500, ProSci) and rabbit anti-MDA5 (1:500, ProSci).
RT-qPCR.
Total RNAs were isolated from different cell lines with or without the indicated PIC treatments. After treatment with DNase I (Ambion, DNA-free™ kit), the cDNA was transcribed with SuperScript II (Invitrogen) with oligo (dT) or random hexamers. Primers for qPCR used: actin-S: GGAC TTC GAG CAA GAG ATG G, actin-AS: AGC ACT GTG TTG GCG TAC AG; eya4-S: CTG TGG AGG TTG CTG GTA CA, eya4-AS: CAA ACC TCA GGG CTG AGA AG; hcgβ-S: TGG CCT TGT CTA CCT CTT GC, hcgβ-AS: GCC TCG TGT ACC TGG CTT TA; hIFNβ-S1: CAT TAC CTG AAG GCC AAG GA, hIFNβ-AS1: CAA TTG TCC AGT CCC AGA GG; hIFNβ-S2: CAG CAG TTC CAG AAG GAG GA, hIFNβ-AS2: AGC CAG GAG GTT CTC AAC AA; lin28-S: GGG GAA TCA CCC TAC AAC CT, lin28-AS: CTT GGC TCC ATG AAT CTG GT; mda5-S: GTC TGG GGC ATG GAG AAT AA, mda5-AS: TGC CCA TGT TGC TGT TAT G; mita-S: TCA AGG ATC GGG TTT ACA GC, mita-AS: CAG AAG AGT TTG GCC TGC TC; pkr-S: CTG TTG ATG GCA CTC TGG AA, pkr-AS: GGT CAA TTG TGG GCT TCA CT; rig-i-S: CTC CCG GCA CAG AAG TGT AT, rig-i-AS: CTT CCT CTG CCT CTG GTT TG; tlr3-S: ACC CAT ACC AAC ATC CCT GA, tlr3-AS: AAG AGT TCA AAG GGG GCA CT; trif-S: TCC TCC TCC TCC TCC TTC AT, trif-AS: GCG TGG AGG ATC ACA AAG TT. Primers for oct3/4 and sox2 were described previously.55 The mRNA expression was quantified relative to actin mRNA in each cell line.
Solexa sequencing and analysis.
RNA-seq libraries were prepared with the mRNA-Seq Sample Prep Kits (Illumina, P/N 1004814) according to the manufacturer's instructions and Solexa deep-sequencing was performed at the UCHC Translational Genomics Core by use of Illumina Genome Analyzer (Illumina). Reads of 75-nt long were aligned to the human genome and junction index by using bowtie87 allowing up to 2 mismatches. Only unique alignments were reported. Gene expression levels are reported here simply as the number of reads that mapped to the annotated exons of each gene using an wiggle-integral program (Duff MO, unpublished).
Acknowledgements
We thank B.R. Graveley and M.O. Duff from the UCHC Translational Genomics Core for helping with the analysis of the Solexa data and members of the lab for helpful comments on the manuscript and throughout the work. Special thanks to H. Zeng for the iPS cell lines from the CT Stem Cell Core Facility. H9 and H14 cells were obtained from the WiCell Research Institute and the CT Stem Cell Core Facility. This work was supported by grant CA04382 from the National Cancer Institute and awards from the State of Connecticut under the Connecticut Stem Cell Research Grants Program to L.L.C. and G.G.C. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the State of Connecticut, the Department of Public Health of the State of Connecticut or Connecticut Innovations, Inc.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/12792
Author Contributions
L.-L. C.: Conception and design, financial support, collection of data, data analysis and interpretation, manuscript writing, approval of manuscript.
L. Y.: Collection of data, data analysis and interpretation, manuscript writing, approval of manuscript.
G. C.: Conception and design, financial support, collection of data, data analysis and interpretation, manuscript writing, approval of manuscript.
Supplementary Material
References
- 1.Wang Q, Carmichael GG. Effects of length and location on the cellular response to double-stranded RNA. Microbiol Mol Biol Rev. 2004;68:432–452. doi: 10.1128/MMBR.68.3.432-452.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar M, Carmichael GG. Antisense RNA: Function and fate of duplex RNA in cells of higher eukaryotes. Microbiol Mol Biol Rev. 1998;62:1415–1434. doi: 10.1128/mmbr.62.4.1415-1434.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baulcombe D. RNA silencing. Trends Biochem Sci. 2005;30:290–293. doi: 10.1016/j.tibs.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 4.Filipowicz W. RNAi: The nuts and bolts of the RISC machine. Cell. 2005;122:17–20. doi: 10.1016/j.cell.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 5.Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsumoto M, Funami K, Oshiumi H, Seya T. Toll-like receptor 3: A link between toll-like receptor, interferon and viruses. Microbiol Immunol. 2004;48:147–154. doi: 10.1111/j.1348-0421.2004.tb03500.x. [DOI] [PubMed] [Google Scholar]
- 7.Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006;442:39–44. doi: 10.1038/nature04946. [DOI] [PubMed] [Google Scholar]
- 8.Gantier MP, Williams BR. The response of mammalian cells to double-stranded RNA. Cytokine Growth Factor Rev. 2007;18:363–371. doi: 10.1016/j.cytogfr.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uematsu S, Akira S. Toll-like receptors and type I interferons. J Biol Chem. 2007;282:15319–15324. doi: 10.1074/jbc.R700009200. [DOI] [PubMed] [Google Scholar]
- 10.Vercammen E, Staal J, Beyaert R. Sensing of viral infection and activation of innate immunity by toll-like receptor 3. Clin Microbiol Rev. 2008;21:13–25. doi: 10.1128/CMR.00022-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kenny EF, O'Neill LAJ. Signalling adaptors used by Toll-like receptors: An update. Cytokine. 2008;43:342–349. doi: 10.1016/j.cyto.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 12.Seya T, Matsumoto M, Ebihara T, Oshiumi H. Functional evolution of the TICAM-1 pathway for extrinsic RNA sensing. Immunol Rev. 2009;227:44–53. doi: 10.1111/j.1600-065X.2008.00723.x. [DOI] [PubMed] [Google Scholar]
- 13.Nakhaei P, Genin P, Civas A, Hiscott J. RIG-I-like receptors: Sensing and responding to RNA virus infection. Semin Immunol. 2009;21:215–222. doi: 10.1016/j.smim.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 14.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 15.McAllister CS, Samuel CE. The RNA-activated protein kinase enhances the induction of interferon-beta and apoptosis mediated by cytoplasmic RNA sensors. J Biol Chem. 2009;284:1644–1651. doi: 10.1074/jbc.M807888200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McAllister CS, Toth AM, Zhang P, Devaux P, Cattaneo R, Samuel CE. Mechanisms of protein kinase PKR-mediated amplification of beta interferon induction by C protein-deficient measles virus. J Virol. 2010;84:380–386. doi: 10.1128/JVI.02630-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lemaire PA, Anderson E, Lary J, Cole JL. Mechanism of PKR activation by dsRNA. J Mol Biol. 2008;381:351–360. doi: 10.1016/j.jmb.2008.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samuel CE. Mechanism of interferon action: Phosphorylation of protein synthesis initiation factor eIF-2 in interferon-treated human cells by a ribosome-associated kinase processing site specificity similar to hemin-regulated rabbit reticulocyte kinase. Proc Natl Acad Sci USA. 1979;76:600–604. doi: 10.1073/pnas.76.2.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar A, Haque J, Lacoste J, Hiscott J, Williams BR. Double-stranded RNA-dependent protein kinase activates transcription factor NFkappaB by phosphorylating IkappaB. Proc Natl Acad Sci USA. 1994;91:6288–6292. doi: 10.1073/pnas.91.14.6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thanos D, Maniatis T. NFkappaB: A lesson in family values. Cell. 1995;80:529–532. doi: 10.1016/0092-8674(95)90506-5. [DOI] [PubMed] [Google Scholar]
- 21.Kumar A, Yang YL, Flati V, Der S, Kadereit S, Deb A, et al. Deficient cytokine signaling in mouse embryo fibroblasts with a targeted deletion in the PKR gene: Role of IRF-1 and NFkappaB. EMBO J. 1997;16:406–416. doi: 10.1093/emboj/16.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams BR. Signal integration via PKR. Sci STKE. 2001:2. doi: 10.1126/stke.2001.89.re2. [DOI] [PubMed] [Google Scholar]
- 23.Kerr IM, Brown RE. pppA2′p5′A2′p5′A: An inhibitor of protein synthesis synthesized with an enzyme fraction from interferon-treated cells. Proc Natl Acad Sci USA. 1978;75:256–260. doi: 10.1073/pnas.75.1.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minks MA, West DK, Benvin S, Greene JJ, Ts'o PO, Baglioni C. Activation of 2′,5′-oligo(A) polymerase and protein kinase of interferon-treated HeLa cells by 2′-O-methylated poly (inosinic acid). Poly(cytidylic acid), Correlations with interferon-inducing activity. J Biol Chem. 1980;255:6403–6407. [PubMed] [Google Scholar]
- 25.Dong B, Silverman RH. 2-5A-dependent RNase molecules dimerize during activation by 2-5A. J Biol Chem. 1995;270:4133–4137. doi: 10.1074/jbc.270.8.4133. [DOI] [PubMed] [Google Scholar]
- 26.Li XL, Blackford JA, Hassel BA. RNase L mediates the antiviral effect of interferon through a selective reduction in viral RNA during encephalomyocarditis virus infection. J Virol. 1998;72:2752–2759. doi: 10.1128/jvi.72.4.2752-2759.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iordanov MS, Paranjape JM, Zhou A, Wong J, Williams BR, Meurs EF, et al. Activation of p38 mitogen-activated protein kinase and c-Jun NH(2)-terminal kinase by double-stranded RNA and encephalomyocarditis virus: involvement of RNase L, protein kinase R and alternative pathways. Mol Cell Biol. 2000;20:617–627. doi: 10.1128/mcb.20.2.617-627.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsumoto M, Funami K, Tanabe M, Oshiumi H, Shingai M, Seto Y, et al. Subcellular localization of Toll-like receptor 3 in human dendritic cells. J Immunol. 2003;171:3154–3162. doi: 10.4049/jimmunol.171.6.3154. [DOI] [PubMed] [Google Scholar]
- 29.Itoh K, Watanabe A, Funami K, Seya T, Matsumoto M. The clathrin-mediated endocytic pathway participates in dsRNA-induced IFNbeta production. J Immunol. 2008;181:5522–5529. doi: 10.4049/jimmunol.181.8.5522. [DOI] [PubMed] [Google Scholar]
- 30.Matsumoto M, Kikkawa S, Kohase M, Miyake K, Seya T. Establishment of a monoclonal antibody against human Toll-like receptor 3 that blocks double-stranded RNA-mediated signaling. Biochem Biophys Res Commun. 2002;293:1364–1369. doi: 10.1016/S0006-291X(02)00380-7. [DOI] [PubMed] [Google Scholar]
- 31.Oshiumi H, Matsumoto M, Funami K, Akazawa T, Seya T. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat Immunol. 2003;4:161–167. doi: 10.1038/ni886. [DOI] [PubMed] [Google Scholar]
- 32.Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, et al. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 33.Jiang Z, Mak TW, Sen G, Li X. Toll-like receptor 3-mediated activation of NFkappaB and IRF3 diverges at Toll-IL-1 receptor domain-containing adapter inducing IFNbeta. Proc Natl Acad Sci USA. 2004;101:3533–3538. doi: 10.1073/pnas.0308496101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meylan E, Burns K, Hofmann K, Blanchetau V, Martinon F, Kelliher M, et al. RIP1 is an essential meidator of Toll-like receptor 3-induced NFkappaB activation. Nat Immunol. 2004;5:503–507. doi: 10.1038/ni1061. [DOI] [PubMed] [Google Scholar]
- 35.Matsumoto M, Seya T. TLR3: Interferon induction by double-stranded RNA including poly(I:C) Adv Drug Deliv Rev. 2008;60:805–812. doi: 10.1016/j.addr.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H, et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 37.Takada E, Okahira S, Sasai M, Funami K, Seya T, Matsumoto M. C-terminal LRRs of human Toll-like receptor 3 control receptor dimerization and signal transmission. Mol Immunol. 2007;44:3633–3640. doi: 10.1016/j.molimm.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 38.Leonard JN, Ghirlando R, Askins J, Bell JK, Margulies DH, Davies DR, et al. The TLR3 signaling complex forms by cooperative receptor dimerization. Proc Natl Acad Sci USA. 2008;105:258–263. doi: 10.1073/pnas.0710779105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 40.Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 41.Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 42.Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baum A, Garcia-Sastre A. Induction of type I interferon by RNA viruses: Cellular receptors and their substrates. Amino Acids. 2009 doi: 10.1007/s00726-009-0374-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 45.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 47.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NFkappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 48.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 49.Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA is an adapter protein required for virus-triggered IFNbeta signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 50.Kumar H, Kawai T, Kato H, Sato S, Takahashi K, Coban C, et al. Essential role of IPS-1 in innate immune responses against RNA viruses. J Exp Med. 2006;203:1795–1803. doi: 10.1084/jem.20060792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun Q, Sun L, Liu HH, Chen X, Seth RB, Forman J, et al. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 2006;24:633–642. doi: 10.1016/j.immuni.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 52.Okabe Y, Sano T, Nagata S. Regulation of the innate immune response by threonine-phosphatase of Eyes absent. Nature. 2009;460:520–524. doi: 10.1038/nature08138. [DOI] [PubMed] [Google Scholar]
- 53.Johnsen IB, Nguyen TT, Ringdal M, Tryggestad AM, Bakke O, Lien E, et al. Toll-like receptor 3 associates with c-Src tyrosine kinase on endosomes to initiate antiviral signaling. EMBO J. 2006;25:3335–3346. doi: 10.1038/sj.emboj.7601222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Funami K, Sasai M, Ohba Y, Oshiumi H, Seya T, Matsumoto M. Spatiotemporal mobilization of Toll/IL-1 receptor domain-containing adaptor molecule-1 in response to dsRNA. J Immunol. 2007;179:6867–6872. doi: 10.4049/jimmunol.179.10.6867. [DOI] [PubMed] [Google Scholar]
- 55.Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: Functional role of a nuclear noncoding RNA. Mol Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schlee M, Hartmann E, Coch C, Wimmenauer V, Janke M, Barchet W, et al. Approaching the RNA ligand for RIG-I? Immunol Rev. 2009;227:66–74. doi: 10.1111/j.1600-065X.2008.00724.x. [DOI] [PubMed] [Google Scholar]
- 57.Wianny F, Zernicka-Goetz M. Specific interference with gene function by double-stranded RNA in early mouse development. Nat Cell Biol. 2000;2:70–75. doi: 10.1038/35000016. [DOI] [PubMed] [Google Scholar]
- 58.Svoboda P, Stein P, Hayashi H, Schultz RM. Selective reduction of dormant maternal mRNAs in mouse oocytes by RNA interference. Development. 2000;127:4147–4156. doi: 10.1242/dev.127.19.4147. [DOI] [PubMed] [Google Scholar]
- 59.Svoboda P, Stein P, Schultz RM. RNAi in mouse oocytes and preimplantation embryos: effectiveness of hairpin dsRNA. Biochem Biophys Res Commun. 2001;287:1099–1104. doi: 10.1006/bbrc.2001.5707. [DOI] [PubMed] [Google Scholar]
- 60.Billy E, Brondani V, Zhang H, Muller U, Filipowicz W. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc Natl Acad Sci USA. 2001;98:14428–14433. doi: 10.1073/pnas.261562698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang S, Tutton S, Pierce E, Yoon K. Specific double-stranded RNA interference in undifferentiated mouse embryonic stem cells. Mol Cell Biol. 2001;21:7807–7816. doi: 10.1128/MCB.21.22.7807-7816.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thomis DC, Samuel CE. Mechanism of interferon action: characterization of the intermolecular autophosphorylation of PKR, the interferon-inducible, RNA-dependent protein kinase. J Virol. 1995;69:5195–5198. doi: 10.1128/jvi.69.8.5195-5198.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang F, Romano PR, Nagamura-Inoue T, Tian B, Dever TE, Mathews MB, et al. Binding of double-stranded RNA to protein kinase PKR is required for dimerization and promotes critical autophosphorylation events in the activation loop. J Biol Chem. 2001;276:24946–24958. doi: 10.1074/jbc.M102108200. [DOI] [PubMed] [Google Scholar]
- 64.Xu RH, Chen X, Li DS, Li R, Addicks GC, Glennon C, et al. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat Biotechnol. 2002;20:1261–1264. doi: 10.1038/nbt761. [DOI] [PubMed] [Google Scholar]
- 65.Xu RH, Peck RM, Li DS, Feng X, Ludwig T, Thomson JA. Basic FGF and suppression of BMP signaling sustain undifferentiated proliferation of human ES cells. Nat Methods. 2005;2:185–190. doi: 10.1038/nmeth744. [DOI] [PubMed] [Google Scholar]
- 66.Zeng H, Park JW, Guo M, Lin G, Crandall L, Compton T, et al. Lack of ABCG2 expression and side population properties in human pluripotent stem cells. Stem Cells. 2009;27:2435–2445. doi: 10.1002/stem.192. [DOI] [PubMed] [Google Scholar]
- 67.Bonin M, Oberstrass J, Lukacs N, Ewert K, Oesterschulze E, Kassing R, et al. Determination of preferential binding sites for anti-dsRNA antibodies on double-stranded RNA by scanning force microscopy. RNA. 2000;6:563–570. doi: 10.1017/s1355838200992318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Balagopal V, Parker R. Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr Opin Cell Biol. 2009;21:403–408. doi: 10.1016/j.ceb.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009;10:430–436. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- 70.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 71.Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, et al. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci USA. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang Z, Carmichael GG. The fate of dsRNA in the nucleus. A p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell. 2001;106:465–475. doi: 10.1016/s0092-8674(01)00466-4. [DOI] [PubMed] [Google Scholar]
- 73.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 74.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 76.Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Qi J, Yu JY, Shcherbata HR, Mathieu J, Wang AJ, Seal S, et al. microRNAs regulate human embryonic stem cell division. Cell Cycle. 2009;8:3729–3741. doi: 10.4161/cc.8.22.10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shalgi R, Brosh R, Oren M, Pilpel Y, Rotter V. Coupling transcriptional and post-transcriptional miRNA regulation in the control of cell fate. Aging. 2009;1:762–770. doi: 10.18632/aging.100085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Barroso-del Jesus A, Lucena-Aguilar G, Menendez P. The miR-302-367 cluster as a potential stemness regulator in ESCs. Cell Cycle. 2009;8:394–398. doi: 10.4161/cc.8.3.7554. [DOI] [PubMed] [Google Scholar]
- 80.Burke DC, Graham CF, Lehman JM. Appearance of interferon inducibility and sensitivity during differentiation of murine teratocarcinoma cells in vitro. Cell. 1978;13:243–248. doi: 10.1016/0092-8674(78)90193-9. [DOI] [PubMed] [Google Scholar]
- 81.Barlow DP, Randle BJ, Burke DC. Interferon synthesis in the early post-implantation mouse embryo. Differentiation. 1984;27:229–235. doi: 10.1111/j.1432-0436.1984.tb01433.x. [DOI] [PubMed] [Google Scholar]
- 82.Francis MK, Lehman JM. Control of beta-interferon expression in murine embryonal carcinoma F9 cells. Mol Cell Biol. 1989;9:3553–3556. doi: 10.1128/mcb.9.8.3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Harada H, Willison K, Sakakibara J, Miyamoto M, Fujita T, Taniguchi T. Absence of the type I IFN system in EC cells: transcriptional activator (IRF-1) and repressor (IRF-2) genes are developmentally regulated. Cell. 1990;63:303–312. doi: 10.1016/0092-8674(90)90163-9. [DOI] [PubMed] [Google Scholar]
- 84.Krause D, Silverman RH, Jacobsen H, Leisy SA, Dieffenbach CW, Friedman RM. Regulation of ppp(A2′p)nA-dependent RNase levels during interferon treatment and cell differentiation. Eur J Biochem. 1985;146:611–618. doi: 10.1111/j.1432-1033.1985.tb08695.x. [DOI] [PubMed] [Google Scholar]
- 85.Kalvakolanu DV, Sen GC. Differentiation-dependent activation of interferon-stimulated gene factors and transcription factor NFkappaB in mouse embryonal carcinoma cells. Proc Natl Acad Sci USA. 1993;90:3167–3171. doi: 10.1073/pnas.90.8.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 87.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.